Comparison of metagenomic next-generation sequencing using cell-free DNA and whole-cell DNA for the diagnoses of pulmonary infections
- PMID: 36439227
- PMCID: PMC9684712
- DOI: 10.3389/fcimb.2022.1042945
Comparison of metagenomic next-generation sequencing using cell-free DNA and whole-cell DNA for the diagnoses of pulmonary infections
Abstract
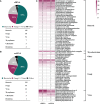
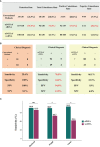
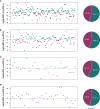
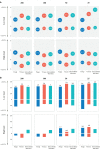
Although the fast-growing metagenomic next-generation sequencing (mNGS) has been used in diagnosing infectious diseases, low detection rate of mNGS in detecting pathogens with low loads limits its extensive application. In this study, 130 patients with suspected pulmonary infections were enrolled, from whom bronchoalveolar lavage fluid (BALF) samples were collected. The conventional tests and mNGS of cell-free DNA (cfDNA) and whole-cell DNA (wcDNA) using BALF were simultaneously performed. mNGS of cfDNA showed higher detection rate (91.5%) and total coincidence rate (73.8%) than mNGS of wcDNA (83.1% and 63.9%) and conventional methods (26.9% and 30.8%). A total of 70 microbes were detected by mNGS of cfDNA, and most of them (60) were also identified by mNGS of wcDNA. The 31.8% (21/66) of fungi, 38.6% (27/70) of viruses, and 26.7% (8/30) of intracellular microbes can be only detected by mNGS of cfDNA, much higher than those [19.7% (13/66), 14.3% (10/70), and 6.7% (2/30)] only detected by mNGS of wcDNA. After in-depth analysis on these microbes with low loads set by reads per million (RPM), we found that more RPM and fungi/viruses/intracellular microbes were detected by mNGS of cfDNA than by mNGS of wcDNA. Besides, the abilities of mNGS using both cfDNA and wcDNA to detect microbes with high loads were similar. We highlighted the advantage of mNGS using cfDNA in detecting fungi, viruses, and intracellular microbes with low loads, and suggested that mNGS of cfDNA could be considered as the first choice for diagnosing pulmonary infections.
Keywords: BALF; MNGs; cell-free DNA; pulmonary infection; whole-cell DNA.
Copyright © 2022 He, Wang, Ke, Zhang, Ning, Zhang, Yang, Shi, Fang, Ming, Li, Zhang, Dong, Liu, Zhou, Xia and Yang.
Conflict of interest statement
JW, JMZ and HX were employed by Hugobiotech. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Figures
Similar articles
-
The value of metagenomic next-generation sequencing with different nucleic acid extracting methods of cell-free DNA or whole-cell DNA in the diagnosis of non-neutropenic pulmonary aspergillosis.Front Cell Infect Microbiol. 2024 Jul 29;14:1398190. doi: 10.3389/fcimb.2024.1398190. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39135636 Free PMC article.
-
Diagnosis of Neurological Infections in Pediatric Patients from Cell-Free DNA Specimens by Using Metagenomic Next-Generation Sequencing.Microbiol Spectr. 2023 Feb 14;11(1):e0253022. doi: 10.1128/spectrum.02530-22. Epub 2023 Jan 18. Microbiol Spectr. 2023. PMID: 36651744 Free PMC article.
-
Comparative analysis of metagenomic next-generation sequencing for pathogenic identification in clinical body fluid samples.BMC Microbiol. 2025 Mar 25;25(1):165. doi: 10.1186/s12866-025-03887-8. BMC Microbiol. 2025. PMID: 40128686 Free PMC article.
-
Metagenomic next generation sequencing of bronchoalveolar lavage fluids for the identification of pathogens in patients with pulmonary infection: A retrospective study.Diagn Microbiol Infect Dis. 2024 Sep;110(1):116402. doi: 10.1016/j.diagmicrobio.2024.116402. Epub 2024 Jun 12. Diagn Microbiol Infect Dis. 2024. PMID: 38878340 Review.
-
The clinical application of metagenomic next-generation sequencing for detecting pathogens in bronchoalveolar lavage fluid: case reports and literature review.Expert Rev Mol Diagn. 2022 May;22(5):575-582. doi: 10.1080/14737159.2022.2071607. Epub 2022 May 2. Expert Rev Mol Diagn. 2022. PMID: 35473493 Review.
Cited by
-
Diagnostic value of DNA or RNA-based metagenomic next-generation sequencing in lower respiratory tract infections.Heliyon. 2024 May 3;10(9):e30712. doi: 10.1016/j.heliyon.2024.e30712. eCollection 2024 May 15. Heliyon. 2024. PMID: 38765131 Free PMC article.
-
Microbial cell-free DNA-sequencing as an addition to conventional diagnostics in neonatal sepsis.Pediatr Res. 2025 Feb;97(2):614-624. doi: 10.1038/s41390-024-03448-1. Epub 2024 Aug 14. Pediatr Res. 2025. PMID: 39143203 Free PMC article.
-
Application of metagenomic next-generation sequencing and targeted metagenomic next-generation sequencing in diagnosing pulmonary infections in immunocompetent and immunocompromised patients.Front Cell Infect Microbiol. 2024 Aug 6;14:1439472. doi: 10.3389/fcimb.2024.1439472. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39165919 Free PMC article.
-
NGS-based Aspergillus detection in plasma and lung lavage of children with invasive pulmonary aspergillosis.NPJ Genom Med. 2025 Mar 17;10(1):24. doi: 10.1038/s41525-025-00482-8. NPJ Genom Med. 2025. PMID: 40097415 Free PMC article.
-
Distribution Patterns of Pathogens Causing Lower Respiratory Tract Infection Based on Metagenomic Next-Generation Sequencing.Infect Drug Resist. 2023 Oct 10;16:6635-6645. doi: 10.2147/IDR.S421383. eCollection 2023. Infect Drug Resist. 2023. PMID: 37840830 Free PMC article.
References
-
- Chen Y., Feng W., Ye K., Guo L., Xia H., Guan Y., et al. . (2021). Application of metagenomic next-generation sequencing in the diagnosis of pulmonary infectious pathogens from bronchoalveolar lavage samples. Front. Cell Infect. Microbiol. 11, 541092. doi: 10.3389/fcimb.2021.541092 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

