Four circadian rhythm-related genes predict incidence and prognosis in hepatocellular carcinoma
- PMID: 36439444
- PMCID: PMC9691441
- DOI: 10.3389/fonc.2022.937403
Four circadian rhythm-related genes predict incidence and prognosis in hepatocellular carcinoma
Abstract
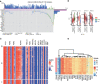
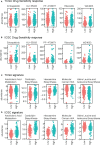
Circadian dysregulation can be involved in the development of malignant tumors, though its relationship with the progression of hepatocellular carcinoma is not yet fully understood. We identified genes related to circadian rhythms from the Cancer Genome Atlas (TCGA), measured gene expression, and conducted genomic difference analysis to construct a circadian rhythm-related signature. The resulting prognosis model proved to be an effective biomarker, as demonstrated by Kaplan-Meier survival analysis for both the training (n = 370, P = 2.687e-10) and external validation cohorts (n = 230, P = 1.45e-02). Further, we found that patients considered 'high risk', with an associated poor prognosis, displayed elevated levels of immune checkpoint genes and immune filtration. We also conducted functional enrichment, which indicated that the risk model showed a significant positive correlation with certain malignant phenotypes, including G2M checkpoint, MYC targets, and the MTORC1 signaling pathway. In summary, we identified a novel circadian rhythm-related signature allowing assessment of prognosis for hepatocellular carcinoma patients, and further can be used to predict immune infiltration sensitivity.
Keywords: Circadian clock; gene signature; immune; liver cancer; overall survival.
Copyright © 2022 Wu, Hu, Zhang, Wang, Li, Qin, Ai, Yi, Wei, Gao and Ouyang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources

