Comparative virulence and antimicrobial resistance distribution of Streptococcus suis isolates obtained from the United States
- PMID: 36439859
- PMCID: PMC9687383
- DOI: 10.3389/fmicb.2022.1043529
Comparative virulence and antimicrobial resistance distribution of Streptococcus suis isolates obtained from the United States
Abstract
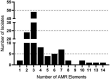
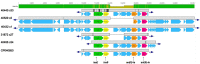
Streptococcus suis is a zoonotic bacterial swine pathogen causing substantial economic and health burdens to the pork industry worldwide. Most S. suis genome sequences available in public databases are from isolates obtained outside the United States. We sequenced the genomes of 106 S. suis isolates from the U.S. and analyzed them to identify their potential to function as zoonotic agents and/or reservoirs for antimicrobial resistance (AMR) dissemination. The objective of this study was to evaluate the genetic diversity of S. suis isolates obtained within the U.S., for the purpose of screening for genomic elements encoding AMR and any factors that could increase or contribute to the capacity of S. suis to transmit, colonize, and/or cause disease in humans. Forty-six sequence types (STs) were identified with ST28 observed as the most prevalent, followed by ST87. Of the 23 different serotypes identified, serotype 2 was the most prevalent, followed by serotype 8 and 3. Of the virulence genes analyzed, the highest nucleotide diversity was observed in sadP, mrp, and ofs. Tetracycline resistance was the most prevalent phenotypic antimicrobial resistance observed followed by macrolide-lincosamide-streptogramin B (MLSB) resistance. Numerous AMR elements were identified, many located within MGE sequences, with the highest frequency observed for ble, tetO and ermB. No genes encoding factors known to contribute to the transmission, colonization, and/or causation of disease in humans were identified in any of the S. suis genomes in this study. This includes the 89 K pathogenicity island carried by the virulent S. suis isolates responsible for human infections. Collectively, the data reported here provide a comprehensive evaluation of the genetic diversity among U.S. S. suis isolates. This study also serves as a baseline for determining any potential risks associated with occupational exposure to these bacteria, while also providing data needed to address public health concerns.
Keywords: Streptococcus suis; antimicrobial resistance; comparative genomics; mobile genetic elements; swine; virulence; whole-genome sequencing.
Copyright © 2022 Nicholson and Bayles.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Aradanas M., Poljak Z., Fittipaldi N., Ricker N., Farzan A. (2021). Serotypes, virulence-associated factors, and antimicrobial resistance of Streptococcus suis isolates recovered from sick and healthy pigs determined by whole-genome sequencing. Front. Vet. Sci. 8:742345. doi: 10.3389/fvets.2021.742345, PMID: - DOI - PMC - PubMed
-
- Carattoli A., Zankari E., Garcia-Fernandez A., Voldby Larsen M., Lund O., Villa L., et al. . (2014). In silico detection and typing of plasmids using plasmid finder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

