A20 binding and inhibitor of nuclear factor kappa B (NF-κB)-1 (ABIN-1): a novel modulator of mitochondrial autophagy
- PMID: 36440857
- PMCID: PMC10191128
- DOI: 10.1152/ajpcell.00493.2022
A20 binding and inhibitor of nuclear factor kappa B (NF-κB)-1 (ABIN-1): a novel modulator of mitochondrial autophagy
Abstract
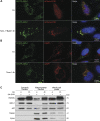
A20 binding inhibitor of nuclear factor kappa B (NF-κB)-1 (ABIN-1), a polyubiquitin-binding protein, is a signal-induced autophagy receptor that attenuates NF-κB-mediated inflammation and cell death. The present study aimed to elucidate the potential role of ABIN-1 in mitophagy, a biological process whose outcome is decisive in diverse physiological and pathological settings. Microtubule-associated proteins 1A/1B light chain 3B-II (LC3B-II) was found to be in complex with ectopically expressed hemagglutinin (HA)-tagged-full length (FL)-ABIN-1. Bacterial expression of ABIN-1 and LC3A and LC3B showed direct binding of ABIN-1 to LC3 proteins, whereas mutations in the LC3-interacting region (LIR) 1 and 2 motifs of ABIN-1 abrogated ABIN-1/LC3B-II complex formation. Importantly, induction of autophagy in HeLa cells resulted in colocalization of ABIN-1 with LC3B-II in autophagosomes and with lysosomal-associated membrane protein 1 (LAMP-1) in autophagolysosomes, leading to degradation of ABIN-1 with p62. Interestingly, ABIN-1 was found to translocate to damaged mitochondria in HeLa-mCherry-Parkin transfected cells. In line with this observation, clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-mediated deletion of ABIN-1 significantly inhibited the degradation of the mitochondrial outer membrane proteins voltage-dependent anion-selective channel 1 (VDAC-1), mitofusin-2 (MFN2), and translocase of outer mitochondrial membrane (TOM)20. In addition, short interfering RNA (siRNA)-mediated knockdown of ABIN-1 significantly decreased lysosomal uptake of mitochondria in HeLa cells expressing mCherry-Parkin and the fluorescence reporter mt-mKEIMA. Collectively, our results identify ABIN-1 as a novel and selective mitochondrial autophagy regulator that promotes mitophagy, thereby adding a new player to the complex cellular machinery regulating mitochondrial homeostasis.
Keywords: LC3-interacting region; mitochondrial outer membrane proteins; mitophagy; selective autophagy receptor.
Conflict of interest statement
Liliana Schaefer is an editor of the
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

