PRDM10 directs FLCN expression in a novel disorder overlapping with Birt-Hogg-Dubé syndrome and familial lipomatosis
- PMID: 36440963
- PMCID: PMC10026250
- DOI: 10.1093/hmg/ddac288
PRDM10 directs FLCN expression in a novel disorder overlapping with Birt-Hogg-Dubé syndrome and familial lipomatosis
Abstract
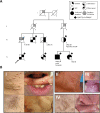
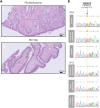
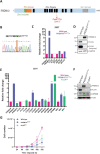
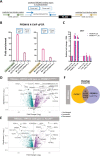
Birt-Hogg-Dubé syndrome (BHD) is an autosomal dominant disorder characterized by fibrofolliculomas, pulmonary cysts, pneumothoraces and renal cell carcinomas. Here, we reveal a novel hereditary disorder in a family with skin and mucosal lesions, extensive lipomatosis and renal cell carcinomas. The proband was initially diagnosed with BHD based on the presence of fibrofolliculomas, but no pathogenic germline variant was detected in FLCN, the gene associated with BHD. By whole exome sequencing we identified a heterozygous missense variant (p.(Cys677Tyr)) in a zinc-finger encoding domain of the PRDM10 gene which co-segregated with the phenotype in the family. We show that PRDM10Cys677Tyr loses affinity for a regulatory binding motif in the FLCN promoter, abrogating cellular FLCN mRNA and protein levels. Overexpressing inducible PRDM10Cys677Tyr in renal epithelial cells altered the transcription of multiple genes, showing overlap but also differences with the effects of knocking out FLCN. We propose that PRDM10 controls an extensive gene program and acts as a critical regulator of FLCN gene transcription in human cells. The germline variant PRDM10Cys677Tyr curtails cellular folliculin expression and underlies a distinguishable syndrome characterized by extensive lipomatosis, fibrofolliculomas and renal cell carcinomas.
© The Author(s) 2022. Published by Oxford University Press.
Figures
Similar articles
-
PRDM10 RCC: A Birt-Hogg-Dubé-like Syndrome Associated With Lipoma and Highly Penetrant, Aggressive Renal Tumors Morphologically Resembling Type 2 Papillary Renal Cell Carcinoma.Urology. 2023 Sep;179:58-70. doi: 10.1016/j.urology.2023.04.035. Epub 2023 Jun 16. Urology. 2023. PMID: 37331486 Free PMC article.
-
Birt-Hogg-Dubé syndrome: novel FLCN frameshift deletion in daughter and father with renal cell carcinomas.Fam Cancer. 2016 Jan;15(1):127-32. doi: 10.1007/s10689-015-9837-5. Fam Cancer. 2016. PMID: 26342594 Free PMC article.
-
Distinctive expression patterns of glycoprotein non-metastatic B and folliculin in renal tumors in patients with Birt-Hogg-Dubé syndrome.Cancer Sci. 2015 Mar;106(3):315-23. doi: 10.1111/cas.12601. Epub 2015 Feb 17. Cancer Sci. 2015. PMID: 25594584 Free PMC article.
-
Pathology of Birt-Hogg-Dubé syndrome: A special reference of pulmonary manifestations in a Japanese population with a comprehensive analysis and review.Pathol Int. 2019 Jan;69(1):1-12. doi: 10.1111/pin.12752. Epub 2019 Jan 11. Pathol Int. 2019. PMID: 30632664 Review.
-
Birt-Hogg-Dubé syndrome: Clinical and molecular aspects of recently identified kidney cancer syndrome.Int J Urol. 2016 Mar;23(3):204-10. doi: 10.1111/iju.13015. Epub 2015 Nov 25. Int J Urol. 2016. PMID: 26608100 Review.
Cited by
-
Recommendations on scuba diving in Birt-Hogg-Dubé syndrome.Expert Rev Respir Med. 2023 Jul-Dec;17(11):1003-1008. doi: 10.1080/17476348.2023.2284375. Epub 2023 Dec 26. Expert Rev Respir Med. 2023. PMID: 37991821 Free PMC article. Review.
-
Maternal PRDM10 activates essential genes for oocyte-to-embryo transition.Nat Commun. 2025 Feb 24;16(1):1939. doi: 10.1038/s41467-025-56991-8. Nat Commun. 2025. PMID: 39994175 Free PMC article.
-
The prevalence of spontaneous pneumothorax in patients with BHD syndrome: a systematic review and meta-analysis.Orphanet J Rare Dis. 2025 May 7;20(1):218. doi: 10.1186/s13023-025-03726-z. Orphanet J Rare Dis. 2025. PMID: 40336059 Free PMC article.
-
PRDM10 RCC: A Birt-Hogg-Dubé-like Syndrome Associated With Lipoma and Highly Penetrant, Aggressive Renal Tumors Morphologically Resembling Type 2 Papillary Renal Cell Carcinoma.Urology. 2023 Sep;179:58-70. doi: 10.1016/j.urology.2023.04.035. Epub 2023 Jun 16. Urology. 2023. PMID: 37331486 Free PMC article.
-
Characteristics, aetiology and implications for management of multiple primary renal tumours: a systematic review.Eur J Hum Genet. 2024 Aug;32(8):887-894. doi: 10.1038/s41431-024-01628-5. Epub 2024 May 27. Eur J Hum Genet. 2024. PMID: 38802529 Free PMC article.
References
-
- Birt, A.R., Hogg, G.R. and Dube, W.J. (1977) Hereditary multiple fibrofolliculomas with trichodiscomas and acrochordons. Arch. Dermatol., 113, 1674–1677. - PubMed
-
- Zbar, B., Alvord, W.G., Glenn, G., Turner, M., Pavlovich, C.P., Schmidt, L., Walther, M., Choyke, P., Weirich, G., Hewitt, S.M. et al. (2002) Risk of renal and colonic neoplasms and spontaneous pneumothorax in the Birt-Hogg-Dube syndrome. Cancer Epidemiol. Biomark. Prev., 11, 393–400. - PubMed
-
- Toro, J.R., Glenn, G., Duray, P., Darling, T., Weirich, G., Zbar, B., Linehan, M. and Turner, M.L. (1999) Birt-Hogg-Dube syndrome: a novel marker of kidney neoplasia. Arch. Dermatol., 135, 1195–1202. - PubMed
-
- Nickerson, M.L., Warren, M.B., Toro, J.R., Matrosova, V., Glenn, G., Turner, M.L., Duray, P., Merino, M., Choyke, P., Pavlovich, C.P. et al. (2002) Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dubé syndrome. Cancer Cell, 2, 157–164. - PubMed
-
- Hudon, V., Sabourin, S., Dydensborg, A.B., Kottis, V., Ghazi, A., Paquet, M., Crosby, K., Pomerleau, V., Uetani, N. and Pause, A. (2010) Renal tumour suppressor function of the Birt-Hogg-Dube syndrome gene product folliculin. J. Med. Genet., 47, 182–189. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

