The multifaceted role of GCM1 during trophoblast differentiation in the human placenta
- PMID: 36442132
- PMCID: PMC9894182
- DOI: 10.1073/pnas.2203071119
The multifaceted role of GCM1 during trophoblast differentiation in the human placenta
Erratum in
-
Correction for Jeyarajah et al., The multifaceted role of GCM1 during trophoblast differentiation in the human placenta.Proc Natl Acad Sci U S A. 2023 Feb 7;120(6):e2300603120. doi: 10.1073/pnas.2300603120. Epub 2023 Feb 2. Proc Natl Acad Sci U S A. 2023. PMID: 36730209 Free PMC article. No abstract available.
Abstract
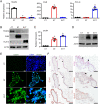
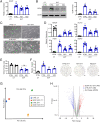
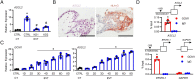
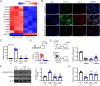
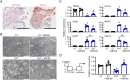
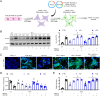
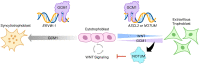
Remodeling of the uterine vasculature by invasive extravillous trophoblasts (EVTs) is a critical aspect of human placentation. Insufficient EVT invasion can lead to severe obstetrical complications like preeclampsia, intrauterine growth restriction, and preterm birth. Glial cells missing-1 (GCM1) is a transcription factor that is crucial for proper placentation in mice, and is highly expressed in human syncytiotrophoblast (ST) and EVTs. GCM1 is classically considered a master regulator of ST formation, but little is known about its contribution to the development and function of EVTs. Therefore, in this study we test the hypothesis that GCM1 is a critical regulator of both EVT and ST development and function. We show that GCM1 is highly expressed in human trophoblast stem (TS) cells differentiated into either ST or EVTs. Knockdown of GCM1 in TS cells hindered differentiation into both ST and EVT pathways. When placed in ST media, GCM1-knockdown cells formed small, unstable clusters; when placed in EVT media, cells had altered morphology and transcript profiles resembling cells trapped in an intermediate state between CT and EVT, and invasive capacity through matrix was reduced. RNA sequencing analysis of GCM1-deficient TS cells revealed downregulation of EVT-associated genes and enrichment in transcripts related to WNT signaling, which was linked to decreased expression of the EVT master regulator ASCL2 and the WNT antagonist NOTUM. Our findings reveal an essential role of GCM1 during ST and EVT development, and suggest that GCM1 regulates differentiation of human TS cells into EVTs by inducing expression of ASCL2 and NOTUM.
Keywords: differentiation; placenta; pregnancy; stem cells; trophoblast.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

