Precision nephrology identified tumor necrosis factor activation variability in minimal change disease and focal segmental glomerulosclerosis
- PMID: 36442540
- PMCID: PMC10347421
- DOI: 10.1016/j.kint.2022.10.023
Precision nephrology identified tumor necrosis factor activation variability in minimal change disease and focal segmental glomerulosclerosis
Abstract
The diagnosis of nephrotic syndrome relies on clinical presentation and descriptive patterns of injury on kidney biopsies, but not specific to underlying pathobiology. Consequently, there are variable rates of progression and response to therapy within diagnoses. Here, an unbiased transcriptomic-driven approach was used to identify molecular pathways which are shared by subgroups of patients with either minimal change disease (MCD) or focal segmental glomerulosclerosis (FSGS). Kidney tissue transcriptomic profile-based clustering identified three patient subgroups with shared molecular signatures across independent, North American, European, and African cohorts. One subgroup had significantly greater disease progression (Hazard Ratio 5.2) which persisted after adjusting for diagnosis and clinical measures (Hazard Ratio 3.8). Inclusion in this subgroup was retained even when clustering was limited to those with less than 25% interstitial fibrosis. The molecular profile of this subgroup was largely consistent with tumor necrosis factor (TNF) pathway activation. Two TNF pathway urine markers were identified, tissue inhibitor of metalloproteinases-1 (TIMP-1) and monocyte chemoattractant protein-1 (MCP-1), that could be used to predict an individual's TNF pathway activation score. Kidney organoids and single-nucleus RNA-sequencing of participant kidney biopsies, validated TNF-dependent increases in pathway activation score, transcript and protein levels of TIMP-1 and MCP-1, in resident kidney cells. Thus, molecular profiling identified a subgroup of patients with either MCD or FSGS who shared kidney TNF pathway activation and poor outcomes. A clinical trial testing targeted therapies in patients selected using urinary markers of TNF pathway activation is ongoing.
Keywords: TNF; data integration; nephrotic syndrome; transcriptomics.
Copyright © 2022 International Society of Nephrology. All rights reserved.
Figures


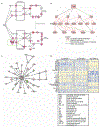
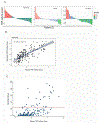

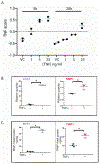

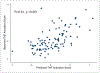
Comment in
-
Precision nephrology: from molecular diagnostics to an individualized therapy.Kidney Int. 2023 Mar;103(3):464-466. doi: 10.1016/j.kint.2022.12.017. Kidney Int. 2023. PMID: 36822751
References
-
- Rovin BH, Adler SG, Barratt J, et al. KDIGO 2021 Clinical Practice Guideline for the Management of Glomerular Diseases. Kidney Int 2021; 100: S1–S276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

