Oxygen-insensitive nitroreductase E. coli NfsA, but not NfsB, is inhibited by fumarate
- PMID: 36443029
- PMCID: PMC10953011
- DOI: 10.1002/prot.26451
Oxygen-insensitive nitroreductase E. coli NfsA, but not NfsB, is inhibited by fumarate
Abstract
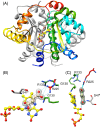
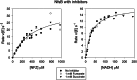
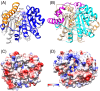
Escherichia coli NfsA and NfsB are founding members of two flavoprotein families that catalyze the oxygen-insensitive reduction of nitroaromatics and quinones by NAD(P)H. This reduction is required for the activity of nitrofuran antibiotics and the enzymes have also been proposed for use with nitroaromatic prodrugs in cancer gene therapy and biocatalysis, but the roles of the proteins in vivo in bacteria are not known. NfsA is NADPH-specific whereas NfsB can also use NADH. The crystal structures of E. coli NfsA and NfsB and several analogs have been determined previously. In our crystal trials, we unexpectedly observed NfsA bound to fumarate. We here present the X-ray structure of the E. coli NfsA-fumarate complex and show that fumarate acts as a weak inhibitor of NfsA but not of NfsB. The structural basis of this differential inhibition is conserved in the two protein families and occurs at fumarate concentrations found in vivo, so impacting the efficacy of these proteins.
Keywords: FMN; Nitroreductase; flavoprotein; fumarate; nitrofuran; prodrug.
© 2022 The Authors. Proteins: Structure, Function, and Bioinformatics published by Wiley Periodicals LLC.
Conflict of interest statement
No commercial interest.
Figures


References
-
- Roldan MD, Perez‐Reinado E, Castillo F, Moreno‐Vivian C. Reduction of polynitroaromatic compounds: the bacterial nitroreductases. FEMS Microbiol Rev. 2008;32(3):474‐500. - PubMed
-
- Williams EM, Little RF, Mowday AM, et al. Nitroreductase gene‐directed enzyme prodrug therapy: insights and advances toward clinical utility. Biochem J. 2015;471:131‐153. - PubMed
-
- Clark AJ, Iwobi M, Cui W, et al. Selective cell ablation in transgenic mice expressing E. coli nitroreductase. Gene Ther. 1997;4:101‐110. - PubMed
-
- Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, Stainier DYR. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236(4):1025‐1035. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

