The fate of early perichondrial cells in developing bones
- PMID: 36443296
- PMCID: PMC9705540
- DOI: 10.1038/s41467-022-34804-6
The fate of early perichondrial cells in developing bones
Abstract
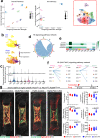
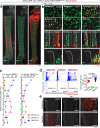
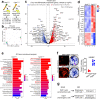
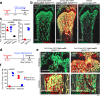
In endochondral bone development, bone-forming osteoblasts and bone marrow stromal cells have dual origins in the fetal cartilage and its surrounding perichondrium. However, how early perichondrial cells distinctively contribute to developing bones remain unidentified. Here we show using in vivo cell-lineage analyses that Dlx5+ fetal perichondrial cells marked by Dlx5-creER do not generate cartilage but sustainably contribute to cortical bone and marrow stromal compartments in a manner complementary to fetal chondrocyte derivatives under the regulation of Hedgehog signaling. Postnatally, Dlx5+ fetal perichondrial cell derivatives preferentially populate the diaphyseal marrow stroma with a dormant adipocyte-biased state and are refractory to parathyroid hormone-induced bone anabolism. Therefore, early perichondrial cells of the fetal cartilage are destined to become an adipogenic subset of stromal cells in postnatal diaphyseal bone marrow, supporting the theory that the adult bone marrow stromal compartments are developmentally prescribed within the two distinct cells-of-origins of the fetal bone anlage.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

