Inactivation of LATS1/2 drives luminal-basal plasticity to initiate basal-like mammary carcinomas
- PMID: 36443313
- PMCID: PMC9705439
- DOI: 10.1038/s41467-022-34864-8
Inactivation of LATS1/2 drives luminal-basal plasticity to initiate basal-like mammary carcinomas
Abstract
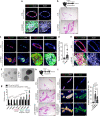
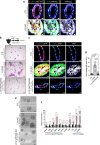
Basal-like breast cancers, an aggressive breast cancer subtype that has poor treatment options, are thought to arise from luminal mammary epithelial cells that undergo basal plasticity through poorly understood mechanisms. Using genetic mouse models and ex vivo primary organoid cultures, we show that conditional co-deletion of the LATS1 and LATS2 kinases, key effectors of Hippo pathway signaling, in mature mammary luminal epithelial cells promotes the development of Krt14 and Sox9-expressing basal-like carcinomas that metastasize over time. Genetic co-deletion experiments revealed that phenotypes resulting from the loss of LATS1/2 activity are dependent on the transcriptional regulators YAP/TAZ. Gene expression analyses of LATS1/2-deleted mammary epithelial cells notably revealed a transcriptional program that associates with human basal-like breast cancers. Our study demonstrates in vivo roles for the LATS1/2 kinases in mammary epithelial homeostasis and luminal-basal fate control and implicates signaling networks induced upon the loss of LATS1/2 activity in the development of basal-like breast cancer.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

