Gene activation guided by nascent RNA-bound transcription factors
- PMID: 36443367
- PMCID: PMC9705438
- DOI: 10.1038/s41467-022-35041-7
Gene activation guided by nascent RNA-bound transcription factors
Abstract
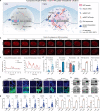
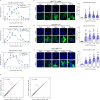
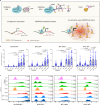
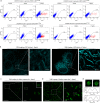
Technologies for gene activation are valuable tools for the study of gene functions and have a wide range of potential applications in bioengineering and medicine. In contrast to existing methods based on recruiting transcriptional modulators via DNA-binding proteins, we developed a strategy termed Narta (nascent RNA-guided transcriptional activation) to achieve gene activation by recruiting artificial transcription factors (aTFs) to transcription sites through nascent RNAs of the target gene. Using Narta, we demonstrate robust activation of a broad range of exogenous and endogenous genes in various cell types, including zebrafish embryos, mouse and human cells. Importantly, the activation is reversible, tunable and specific. Moreover, Narta provides better activation potency of some expressed genes than CRISPRa and, when used in combination with CRISPRa, has an enhancing effect on gene activation. Quantitative imaging illustrated that nascent RNA-directed aTFs could induce the high-density assembly of coactivators at transcription sites, which may explain the larger transcriptional burst size induced by Narta. Overall, our work expands the gene activation toolbox for biomedical research.
© 2022. The Author(s).
Conflict of interest statement
Y.L., W.Z., and B.C. submitted a patent application (Chinese Patent Application No. 2022111029154) on the design and application of a nascent RNA-guided gene activation method based on the technology developed in this paper. All other authors declare no competing interests.
Figures

Similar articles
-
Imaging nascent transcription in wholemount vertebrate embryos to characterize zygotic genome activation.Methods Enzymol. 2020;638:139-165. doi: 10.1016/bs.mie.2020.03.002. Epub 2020 Mar 23. Methods Enzymol. 2020. PMID: 32416911 Free PMC article.
-
A highly potent artificial transcription factor.Biochemistry. 2002 Jun 11;41(23):7209-16. doi: 10.1021/bi015906b. Biochemistry. 2002. PMID: 12044151
-
Inhibition of endogenous MTF-1 signaling in zebrafish embryos identifies novel roles for MTF-1 in development.Biochim Biophys Acta. 2014 Sep;1843(9):1818-33. doi: 10.1016/j.bbamcr.2014.04.015. Epub 2014 Apr 18. Biochim Biophys Acta. 2014. PMID: 24751692 Free PMC article.
-
Molecular genetics of the RNA polymerase II general transcriptional machinery.Microbiol Mol Biol Rev. 1998 Jun;62(2):465-503. doi: 10.1128/MMBR.62.2.465-503.1998. Microbiol Mol Biol Rev. 1998. PMID: 9618449 Free PMC article. Review.
-
Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation.Mol Endocrinol. 2003 Oct;17(10):1901-9. doi: 10.1210/me.2002-0384. Epub 2003 Jul 31. Mol Endocrinol. 2003. PMID: 12893880 Review.
Cited by
-
Dynamic properties of transcriptional condensates modulate CRISPRa-mediated gene activation.Nat Commun. 2025 Feb 14;16(1):1640. doi: 10.1038/s41467-025-56735-8. Nat Commun. 2025. PMID: 39952932 Free PMC article.
-
Transcription factors and splice factors - interconnected regulators of stem cell differentiation.Curr Stem Cell Rep. 2023 Jun;9(2):31-41. doi: 10.1007/s40778-023-00227-2. Epub 2023 Jun 29. Curr Stem Cell Rep. 2023. PMID: 38939410 Free PMC article.
-
Histone lactylation mediated by Fam172a in POMC neurons regulates energy balance.Nat Commun. 2024 Nov 22;15(1):10111. doi: 10.1038/s41467-024-54488-4. Nat Commun. 2024. PMID: 39578459 Free PMC article.
-
Stochastic Modeling of Biophysical Responses to Perturbation.bioRxiv [Preprint]. 2024 Jul 5:2024.07.04.602131. doi: 10.1101/2024.07.04.602131. bioRxiv. 2024. PMID: 39005347 Free PMC article. Preprint.
-
RGS1 Enhancer RNA Promotes Gene Transcription by Recruiting Transcription Factor FOXJ3 and Facilitates Osteoclastogenesis Through PLC-IP3R-dependent Ca2+ Response in Rheumatoid Arthritis.Inflammation. 2025 Feb;48(1):447-463. doi: 10.1007/s10753-024-02067-6. Epub 2024 Jun 21. Inflammation. 2025. PMID: 38904871
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

