Manipulating chromatin architecture in C. elegans
- PMID: 36443798
- PMCID: PMC9706983
- DOI: 10.1186/s13072-022-00472-5
Manipulating chromatin architecture in C. elegans
Abstract
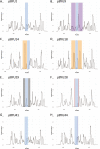
Background: Nucleosome-mediated chromatin compaction has a direct effect on the accessibility of trans-acting activators and repressors to DNA targets and serves as a primary regulatory agent of genetic expression. Understanding the nature and dynamics of chromatin is fundamental to elucidating the mechanisms and factors that epigenetically regulate gene expression. Previous work has shown that there are three types of canonical sequences that strongly regulate nucleosome positioning and thus chromatin accessibility: putative nucleosome-positioning elements, putative nucleosome-repelling sequences, and homopolymeric runs of A/T. It is postulated that these elements can be used to remodel chromatin in C. elegans. Here we show the utility of such elements in vivo, and the extreme efficacy of a newly discovered repelling sequence, PRS-322.
Results: In this work, we show that it is possible to manipulate nucleosome positioning in C. elegans solely using canonical and putative positioning sequences. We have not only tested previously described sequences such as the Widom 601, but also have tested additional nucleosome-positioning sequences: the Trifonov sequence, putative repelling sequence-322 (PRS-322), and various homopolymeric runs of A and T nucleotides.
Conclusions: Using each of these types of putative nucleosome-positioning sequences, we demonstrate their ability to alter the nucleosome profile in C. elegans as evidenced by altered nucleosome occupancy and positioning in vivo. Additionally, we show the effect that PRS-322 has on nucleosome-repelling and chromatin remodeling.
Keywords: C. elegans; Chromatin; Epigenetics; Nucleosome occupancy; Nucleosome positioning; Nucleosomes; PRS-322; Transgene; Widom 601.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

