STING Suppresses Mitochondrial VDAC2 to Govern RCC Growth Independent of Innate Immunity
- PMID: 36445063
- PMCID: PMC9875608
- DOI: 10.1002/advs.202203718
STING Suppresses Mitochondrial VDAC2 to Govern RCC Growth Independent of Innate Immunity
Abstract
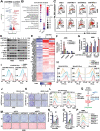
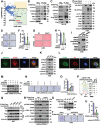
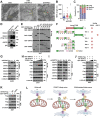
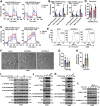
STING is an innate immune sensor for immune surveillance of viral/bacterial infection and maintenance of an immune-friendly microenvironment to prevent tumorigenesis. However, if and how STING exerts innate immunity-independent function remains elusive. Here, the authors report that STING expression is increased in renal cell carcinoma (RCC) patients and governs tumor growth through non-canonical innate immune signaling involving mitochondrial ROS maintenance and calcium homeostasis. Mitochondrial voltage-dependent anion channel VDAC2 is identified as a new STING binding partner. STING depletion potentiates VDAC2/GRP75-mediated MERC (mitochondria-ER contact) formation to increase mitochondrial ROS/calcium levels, impairs mitochondria function, and suppresses mTORC1/S6K signaling leading to RCC growth retardation. STING interaction with VDAC2 occurs through STING-C88/C91 palmitoylation and inhibiting STING palmitoyl-transferases ZDHHCs by 2-BP significantly impedes RCC cell growth alone or in combination with sorafenib. Together, these studies reveal an innate immunity-independent function of STING in regulating mitochondrial function and growth in RCC, providing a rationale to target the STING/VDAC2 interaction in treating RCC.
Keywords: 2-BP; STING; VDAC2; innate immunity-independent; mTORC1; mitochondrial homeostasis.
© 2022 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Akira S., Uematsu S., Takeuchi O., Cell 2006, 124, 783. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
