Oral Bacteriome and Mycobiome across Stages of Oral Carcinogenesis
- PMID: 36445134
- PMCID: PMC9769585
- DOI: 10.1128/spectrum.02737-22
Oral Bacteriome and Mycobiome across Stages of Oral Carcinogenesis
Abstract
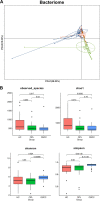
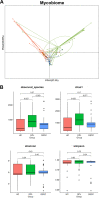
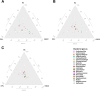
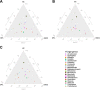
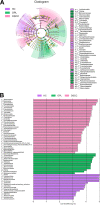
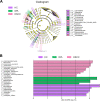
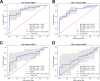
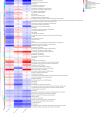
Oral microbial dysbiosis contributes to the development of oral squamous cell carcinoma (OSCC). Numerous studies have focused on variations in the oral bacterial microbiota of patients with OSCC. However, similar studies on fungal microbiota, another integral component of the oral microbiota, are scarce. Moreover, there is an evidence gap regarding the role that microecosystems play in different niches of the oral cavity at different stages of oral carcinogenesis. Here, we catalogued the microbial communities in the human oral cavity by profiling saliva, gingival plaque, and mucosal samples at different stages of oral carcinogenesis. We analyzed the oral bacteriome and mycobiome along the health-premalignancy-carcinoma sequence. Some species, including Prevotella intermedia, Porphyromonas endodontalis, Acremonium exuviarum, and Aspergillus fumigatus, were enriched, whereas others, such as Streptococcus salivarius subsp. salivarius, Scapharca broughtonii, Mortierella echinula, and Morchella septimelata, were depleted in OSCC. These findings suggest that an array of signature species, including bacteria and fungi, are closely associated with oral carcinogenesis. OSCC-associated diversity differences, species distinction, and functional alterations were most remarkable in mucosal samples, not in gingival plaque or saliva samples, suggesting an urgent need to define oral carcinogenesis-associated microbial dysbiosis based on the spatial microbiome. IMPORTANCE Abundant oral microorganisms constitute a complex microecosystem within the oral environment of the host, which plays a critical role in the adjustment of various physiological and pathological states of the oral cavity. In this study, we demonstrated that variations in the "core microbiome" may be used to predict carcinogenesis. In addition, sample data collected from multiple oral sites along the health-premalignancy-carcinoma sequence increase our understanding of the microecosystems of different oral niches and their specific changes during oral carcinogenesis. This work provides insight into the roles of bacteria and fungi in OSCC and may contribute to the development of early diagnostic assays and novel treatments.
Keywords: bacteriome; carcinogenesis; mycobiome; oral cavity; plaque; saliva.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Hill SJ, D'Andrea AD. 2019. Predictive potential of head and neck squamous cell carcinoma organoids. Cancer Discov 9:828–830. doi: 10.1158/2159-8290.CD-19-0527. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

