Aspergillus flavus genetic structure at a turkey farm
- PMID: 36445341
- PMCID: PMC9857098
- DOI: 10.1002/vms3.1015
Aspergillus flavus genetic structure at a turkey farm
Abstract
Background: The ubiquitous environmental fungus Aspergillus flavus is also a life-threatening avian pathogen.
Objectives: This study aimed to assess the genetic diversity and population structure of A. flavus isolated from turkey lung biopsy or environmental samples collected in a poultry farm.
Methods: A. flavus isolates were identified using both morphological and ITS sequence features. Multilocus microsatellite genotyping was performed by using a panel of six microsatellite markers. Population genetic indices were computed using FSTAT and STRUCTURE. A minimum-spanning tree (MST) and UPGMA dendrogram were drawn using BioNumerics and NTSYS-PC, respectively.
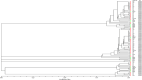
Results: The 63 environmental (air, surfaces, eggshells and food) A. flavus isolates clustered in 36 genotypes (genotypic diversity = 0.57), and the 19 turkey lung biopsies isolates clustered in 17 genotypes (genotypic diversity = 0.89). The genetic structure of environmental and avian A. flavus populations were clearly differentiated, according to both F-statistics and Bayesian model-based analysis' results. The Bayesian approach indicated gene flow between both A. flavus populations. The MST illustrated the genetic structure of this A. flavus population split in nine clusters, including six singletons.
Conclusions: Our results highlight the distinct genetic structure of environmental and avian A. flavus populations, indicative of a genome-based adaptation of isolates involved in avian aspergillosis.
Keywords: Aspergillus flavus; Bayesian model; F-statistics; Meleagris gallopavo; aspergillosis; avian aspergillosis; environment; genetic structure; microsatellite markers; population genetics; typing.
© 2022 The Authors. Veterinary Medicine and Science published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Cafarchia, C. , Camarda, A. , Iatta, R. , Danesi, P. , Favuzzi, V. , Di Paola, G. , Pugliese, N. , Caroli, A. , Montagna, M. T. , & Otranto, D. (2014). Environmental contamination by Aspergillus spp. in laying hen farms and associated health risks for farm workers. Journal of Medical Microbiology, 63(Pt 3), 464–470. - PubMed
-
- Chen, Y. , Litvintseva, A. P. , Frazzitta, A. E. , Haverkamp, M. R. , Wang, L. , Fang, C. , Muthoga, C. , Mitchell, T. G. , & Perfect, J R. (2015). Comparative analyses of clinical and environmental populations of Cryptococcus neoformans in Botswana. Molecular Ecology, 24(14), 3559–3571. - PMC - PubMed
-
- de Hoog, G. S. , Nishikaku, A. S. , Fernandez‐Zeppenfeldt, G. , Padín‐González, C. , Burger, E. , Badali, H. , Richard‐Yegres, N. , & van den Ende, A. H. (2007). Molecular analysis and pathogenicity of the Cladophialophora carrionii complex, with the description of a novel species. Studies in Mycology, 58, 219–234. - PMC - PubMed
-
- Earl, D. A. , & vonHoldt, B. M. (2011). STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4(2), 359–361.
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

