Inducible degradation of the Drosophila Mediator subunit Med19 reveals its role in regulating developmental but not constitutively-expressed genes
- PMID: 36445897
- PMCID: PMC9707739
- DOI: 10.1371/journal.pone.0275613
Inducible degradation of the Drosophila Mediator subunit Med19 reveals its role in regulating developmental but not constitutively-expressed genes
Abstract
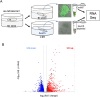
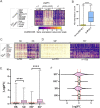
The multi-subunit Mediator complex plays a critical role in gene expression by bridging enhancer-bound transcription factors and the RNA polymerase II machinery. Although experimental case studies suggest differential roles of Mediator subunits, a comprehensive view of the specific set of genes regulated by individual subunits in a developing tissue is still missing. Here we address this fundamental question by focusing on the Med19 subunit and using the Drosophila wing imaginal disc as a developmental model. By coupling auxin-inducible degradation of endogenous Med19 in vivo with RNA-seq, we got access to the early consequences of Med19 elimination on gene expression. Differential gene expression analysis reveals that Med19 is not globally required for mRNA transcription but specifically regulates positively or negatively less than a quarter of the expressed genes. By crossing our transcriptomic data with those of Drosophila gene expression profile database, we found that Med19-dependent genes are highly enriched with spatially-regulated genes while the expression of most constitutively expressed genes is not affected upon Med19 loss. Whereas globally downregulation does not exceed upregulation, we identified a functional class of genes encoding spatially-regulated transcription factors, and more generally developmental regulators, responding unidirectionally to Med19 loss with an expression collapse. Moreover, we show in vivo that the Notch-responsive wingless and the E(spl)-C genes require Med19 for their expression. Combined with experimental evidences suggesting that Med19 could function as a direct transcriptional effector of Notch signaling, our data support a model in which Med19 plays a critical role in the transcriptional activation of developmental genes in response to cell signaling pathways.
Copyright: © 2022 Jullien et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

