The N-Myc-responsive lncRNA MILIP promotes DNA double-strand break repair through non-homologous end joining
- PMID: 36445966
- PMCID: PMC9894261
- DOI: 10.1073/pnas.2208904119
The N-Myc-responsive lncRNA MILIP promotes DNA double-strand break repair through non-homologous end joining
Erratum in
-
Correction for Wang et al., The N-Myc-responsive lncRNA MILIP promotes DNA double-strand break repair through non-homologous end joining.Proc Natl Acad Sci U S A. 2024 Jul 16;121(29):e2412548121. doi: 10.1073/pnas.2412548121. Epub 2024 Jul 8. Proc Natl Acad Sci U S A. 2024. PMID: 38976744 Free PMC article. No abstract available.
Abstract
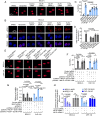
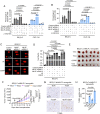
The protooncoprotein N-Myc, which is overexpressed in approximately 25% of neuroblastomas as the consequence of MYCN gene amplification, has long been postulated to regulate DNA double-strand break (DSB) repair in neuroblastoma cells, but experimental evidence of this function is presently scant. Here, we show that N-Myc transcriptionally activates the long noncoding RNA MILIP to promote nonhomologous end-joining (NHEJ) DNA repair through facilitating Ku70-Ku80 heterodimerization in neuroblastoma cells. High MILIP expression was associated with poor outcome and appeared as an independent prognostic factor in neuroblastoma patients. Knockdown of MILIP reduced neuroblastoma cell viability through the induction of apoptosis and inhibition of proliferation, retarded neuroblastoma xenograft growth, and sensitized neuroblastoma cells to DNA-damaging therapeutics. The effect of MILIP knockdown was associated with the accumulation of DNA DSBs in neuroblastoma cells largely due to decreased activity of the NHEJ DNA repair pathway. Mechanistical investigations revealed that binding of MILIP to Ku70 and Ku80 increased their heterodimerization, and this was required for MILIP-mediated promotion of NHEJ DNA repair. Disrupting the interaction between MILIP and Ku70 or Ku80 increased DNA DSBs and reduced cell viability with therapeutic potential revealed where targeting MILIP using Gapmers cooperated with the DNA-damaging drug cisplatin to inhibit neuroblastoma growth in vivo. Collectively, our findings identify MILIP as an N-Myc downstream effector critical for activation of the NHEJ DNA repair pathway in neuroblastoma cells, with practical implications of MILIP targeting, alone and in combination with DNA-damaging therapeutics, for neuroblastoma treatment.
Keywords: DNA repair; MYCN; N-Myc; lncRNA; neuroblastoma.
Conflict of interest statement
The authors declare no competing interest.
Figures




References
-
- Brodeur G. M., Neuroblastoma: Biological insights into a clinical enigma. Nat. Rev. Cancer 3, 203–216 (2003). - PubMed
-
- Liu P. Y., Atmadibrata B., Mondal S., Tee A. E., Liu T., NCYM is upregulated by lncUSMycN and modulates N-Myc expression. Int. J. Oncol. 49, 2464–2470 (2016). - PubMed
-
- Papadopoulos D., et al. , MYCN recruits the nuclear exosome complex to RNA polymerase II to prevent transcription-replication conflicts. Mol. Cell 82, 159–176.e112 (2022). - PubMed
-
- Luoto K. R., et al. , Tumor cell kill by c-MYC depletion: Role of MYC-regulated genes that control DNA double-strand break repair. Cancer Res. 70, 8748–8759 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

