Gabrb3 is required for the functional integration of pyramidal neuron subtypes in the somatosensory cortex
- PMID: 36446382
- PMCID: PMC9852093
- DOI: 10.1016/j.neuron.2022.10.037
Gabrb3 is required for the functional integration of pyramidal neuron subtypes in the somatosensory cortex
Abstract
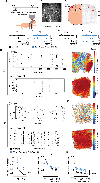
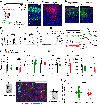
Dysfunction of gamma-aminobutyric acid (GABA)ergic circuits is strongly associated with neurodevelopmental disorders. However, it is unclear how genetic predispositions impact circuit assembly. Using in vivo two-photon and widefield calcium imaging in developing mice, we show that Gabrb3, a gene strongly associated with autism spectrum disorder (ASD) and Angelman syndrome (AS), is enriched in contralaterally projecting pyramidal neurons and is required for inhibitory function. We report that Gabrb3 ablation leads to a developmental decrease in GABAergic synapses, increased local network synchrony, and long-lasting enhancement in functional connectivity of contralateral-but not ipsilateral-pyramidal neuron subtypes. In addition, Gabrb3 deletion leads to increased cortical response to tactile stimulation at neonatal stages. Using human transcriptomics and neuroimaging datasets from ASD subjects, we show that the spatial distribution of GABRB3 expression correlates with atypical connectivity in these subjects. Our studies reveal a requirement for Gabrb3 during the emergence of interhemispheric circuits for sensory processing.
Keywords: Angelman; GABA; GABRB3; callosal projections; corpus callosum; interhemispheric connectivity; network synchrony; projection neurons; pyramidal neurons.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






Comment in
-
aGABRacadabra: A surprising new role for GABAA receptors in cortical development.Neuron. 2023 Jan 18;111(2):146-149. doi: 10.1016/j.neuron.2022.12.019. Neuron. 2023. PMID: 36657397
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

