EZH2-H3K27me3 mediated KRT14 upregulation promotes TNBC peritoneal metastasis
- PMID: 36446780
- PMCID: PMC9708848
- DOI: 10.1038/s41467-022-35059-x
EZH2-H3K27me3 mediated KRT14 upregulation promotes TNBC peritoneal metastasis
Abstract
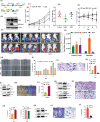
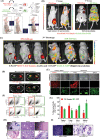
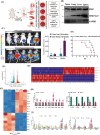
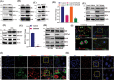
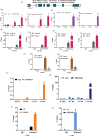
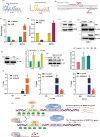
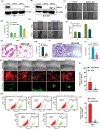
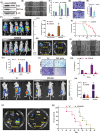
Triple-Negative Breast Cancer (TNBC) has a poor prognosis and adverse clinical outcomes among all breast cancer subtypes as there is no available targeted therapy. Overexpression of Enhancer of zeste homolog 2 (EZH2) has been shown to correlate with TNBC's poor prognosis, but the contribution of EZH2 catalytic (H3K27me3) versus non-catalytic EZH2 (NC-EZH2) function in TNBC progression remains elusive. We reveal that selective hyper-activation of functional EZH2 (H3K27me3) over NC-EZH2 alters TNBC metastatic landscape and fosters its peritoneal metastasis, particularly splenic. Instead of H3K27me3-mediated repression of gene expression; here, it promotes KRT14 transcription by attenuating binding of repressor SP1 to its promoter. Further, KRT14 loss significantly reduces TNBC migration, invasion, and peritoneal metastasis. Consistently, human TNBC metastasis displays positive correlation between H3K27me3 and KRT14 levels. Finally, EZH2 knockdown or H3K27me3 inhibition by EPZ6438 reduces TNBC peritoneal metastasis. Altogether, our preclinical findings suggest a rationale for targeting TNBC with EZH2 inhibitors.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Wolff AC, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch. Pathol. Lab. Med. 2014;138:241–256. doi: 10.5858/arpa.2013-0953-SA. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

