CARD11 mutation and HBZ expression induce lymphoproliferative disease and adult T-cell leukemia/lymphoma
- PMID: 36446869
- PMCID: PMC9709164
- DOI: 10.1038/s42003-022-04284-x
CARD11 mutation and HBZ expression induce lymphoproliferative disease and adult T-cell leukemia/lymphoma
Abstract
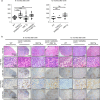
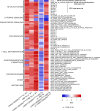
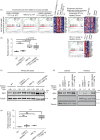
Adult T-cell leukemia/lymphoma (ATL) is caused by human T-cell leukemia virus type 1 (HTLV-1). In addition to HTLV-1 bZIP factor (HBZ), a leukemogenic antisense transcript of HTLV-1, abnormalities of genes involved in TCR-NF-κB signaling, such as CARD11, are detected in about 90% of patients. Utilizing mice expressing CD4+ T cell-specific CARD11(E626K) and/or CD4+ T cell-specific HBZ, namely CARD11(E626K)CD4-Cre mice, HBZ transgenic (Tg) mice, and CARD11(E626K)CD4-Cre;HBZ Tg double transgenic mice, we clarify these genes' pathogenetic effects. CARD11(E626K)CD4-Cre and HBZ Tg mice exhibit lymphocytic invasion to many organs, including the lungs, and double transgenic mice develop lymphoproliferative disease and increase CD4+ T cells in vivo. CARD11(E626K) and HBZ cooperatively activate the non-canonical NF-κB pathway, IRF4 targets, BATF3/IRF4/HBZ transcriptional network, MYC targets, and E2F targets. Most KEGG and HALLMARK gene sets enriched in acute-type ATL are also enriched in double transgenic mice, indicating that these genes cooperatively contribute to ATL development.
© 2022. The Author(s).
Conflict of interest statement
K. Shimoda has received consulting fees from Novartis Pharma, Takeda Pharmaceutical, Bristol-Myers, Shire Japan, and Celgene, all outside the submitted work, and has received research grants from Perseus Proteomics, Pharma Essentia Japan KK, AbbVie GK, Astellas Pharma, MSD, Chugai Pharmaceutical, Kyowa Kirin, Pfizer, Novartis Pharma, Otsuka Pharmaceutical, Asahi Kasei Medical, all outside the submitted work. K.K. holds stock in Asahi Genomics, has a patent for genetic alterations as a biomarker in T-cell lymphomas, and has received research funding from Chordia Therapeutics outside the submitted work. The remaining authors declare no competing interests.
Figures






References
-
- WHO Classification of Tumors of Haematopoietic and Lymphoid Tissues. Adult T-cell leukaemia/lymphoma (ed. Swerdlow, S. H. et al.). 281–284 (IARC press Lyon, 2008).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

