Electrocardiogram lead conversion from single-lead blindly-segmented signals
- PMID: 36447207
- PMCID: PMC9710059
- DOI: 10.1186/s12911-022-02063-6
Electrocardiogram lead conversion from single-lead blindly-segmented signals
Abstract
Background: The standard configuration's set of twelve electrocardiogram (ECG) leads is optimal for the medical diagnosis of diverse cardiac conditions. However, it requires ten electrodes on the patient's limbs and chest, which is uncomfortable and cumbersome. Interlead conversion methods can reconstruct missing leads and enable more comfortable acquisitions, including in wearable devices, while still allowing for adequate diagnoses. Currently, methodologies for interlead ECG conversion either require multiple reference (input) leads and/or require input signals to be temporally aligned considering the ECG landmarks.
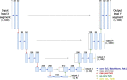
Methods: Unlike the methods in the literature, this paper studies the possibility of converting ECG signals into all twelve standard configuration leads using signal segments from only one reference lead, without temporal alignment (blindly-segmented). The proposed methodology is based on a deep learning encoder-decoder U-Net architecture, which is compared with adaptations based on convolutional autoencoders and label refinement networks. Moreover, the method is explored for conversion with one single shared encoder or multiple individual encoders for each lead.
Results: Despite the more challenging settings, the proposed methodology was able to attain state-of-the-art level performance in multiple target leads, and both lead I and lead II seem especially suitable to convert certain sets of leads. In cross-database tests, the methodology offered promising results despite acquisition setup differences. Furthermore, results show that the presence of medical conditions does not have a considerable effect on the method's performance.
Conclusions: This study shows the feasibility of converting ECG signals using single-lead blindly-segmented inputs. Although the results are promising, further efforts should be devoted towards the improvement of the methodologies, especially the robustness to diverse acquisition setups, in order to be applicable to cardiac health monitoring in wearable devices and less obtrusive clinical scenarios.
Keywords: Autoencoder; Conversion; Deep learning; Electrocardiogram (ECG); Leads; U-Net.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
-
- Pinto JR, Cardoso JS, Lourenço A. Evolution, current challenges, and future possibilities in ECG biometrics. IEEE Access. 2018;6:34746–34776. doi: 10.1109/ACCESS.2018.2849870. - DOI
-
- Matyschik M, Mauranen H, Bonizzi P, Karel J. Feasibility of ECG reconstruction from minimal lead sets using convolutional neural networks. In: Computing in Cardiology 2020.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

