Image analysis workflows to reveal the spatial organization of cell nuclei and chromosomes
- PMID: 36447428
- PMCID: PMC9754023
- DOI: 10.1080/19491034.2022.2144013
Image analysis workflows to reveal the spatial organization of cell nuclei and chromosomes
Abstract
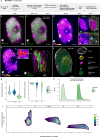
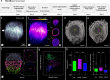
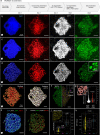
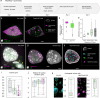
Nucleus, chromatin, and chromosome organization studies heavily rely on fluorescence microscopy imaging to elucidate the distribution and abundance of structural and regulatory components. Three-dimensional (3D) image stacks are a source of quantitative data on signal intensity level and distribution and on the type and shape of distribution patterns in space. Their analysis can lead to novel insights that are otherwise missed in qualitative-only analyses. Quantitative image analysis requires specific software and workflows for image rendering, processing, segmentation, setting measurement points and reference frames and exporting target data before further numerical processing and plotting. These tasks often call for the development of customized computational scripts and require an expertise that is not broadly available to the community of experimental biologists. Yet, the increasing accessibility of high- and super-resolution imaging methods fuels the demand for user-friendly image analysis workflows. Here, we provide a compendium of strategies developed by participants of a training school from the COST action INDEPTH to analyze the spatial distribution of nuclear and chromosomal signals from 3D image stacks, acquired by diffraction-limited confocal microscopy and super-resolution microscopy methods (SIM and STED). While the examples make use of one specific commercial software package, the workflows can easily be adapted to concurrent commercial and open-source software. The aim is to encourage biologists lacking custom-script-based expertise to venture into quantitative image analysis and to better exploit the discovery potential of their images.Abbreviations: 3D FISH: three-dimensional fluorescence in situ hybridization; 3D: three-dimensional; ASY1: ASYNAPTIC 1; CC: chromocenters; CO: Crossover; DAPI: 4',6-diamidino-2-phenylindole; DMC1: DNA MEIOTIC RECOMBINASE 1; DSB: Double-Strand Break; FISH: fluorescence in situ hybridization; GFP: GREEN FLUORESCENT PROTEIN; HEI10: HUMAN ENHANCER OF INVASION 10; NCO: Non-Crossover; NE: Nuclear Envelope; Oligo-FISH: oligonucleotide fluorescence in situ hybridization; RNPII: RNA Polymerase II; SC: Synaptonemal Complex; SIM: structured illumination microscopy; ZMM (ZIP: MSH4: MSH5 and MER3 proteins); ZYP1: ZIPPER-LIKE PROTEIN 1.
Keywords: 3D organization; Nucleus; RNA Pol II; SIM; STED imaging; chromatin; chromosome; crossovers; image analysis; meiosis; metaphase; mitosis; nuclear bodies; nuclear speckles; oligo FISH; pachytene; quantification; segmentation; spatial distribution; transcription factories.
Conflict of interest statement
Author contribution
The work was conceived by CB; carried out and conceptually refined by CB, RSR, DR, CJ, AN, KK, MK, MAA, VS, CT, IC, K, MP, IK, AS, SM, and SD; supervised by CB, CT, VS, ML, SH, RS, DS, and AP; and written by CB, RSR, CJ, AN, KK, MK, MAA, VS, CT, and IC. All authors read and revised the manuscript.
Figures

References
-
- Jerkovic I, Cavalli G.. Understanding 3D genome organization by multidisciplinary methods. Nat Rev Mol Cell Biol. 2021;22:511–528. - PubMed
-
- Dar AS, Padha D. Medical image segmentation: a review of recent techniques, advancements and a comprehensive comparison. Int J Comput Sci Eng. 2019;7:114–124.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
