The Sia System and c-di-GMP Play a Crucial Role in Controlling Cell-Association of Psl in Planktonic P. aeruginosa
- PMID: 36448788
- PMCID: PMC9794950
- DOI: 10.1128/jb.00335-22
The Sia System and c-di-GMP Play a Crucial Role in Controlling Cell-Association of Psl in Planktonic P. aeruginosa
Abstract
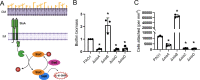
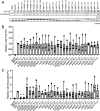
Many bacterial species use the secondary messenger, c-di-GMP, to promote the production of biofilm matrix components. In Pseudomonas aeruginosa, c-di-GMP production is stimulated upon initial surface contact and generally remains high throughout biofilm growth. Transcription of several gene clusters, including the Sia signal transduction system, are induced in response to high cellular levels of c-di-GMP. The output of this system is SiaD, a diguanylate cyclase whose activity is induced in the presence of the detergent SDS. Previous studies demonstrated that Sia-mediated cellular aggregation is a key feature of P. aeruginosa growth in the presence of SDS. Here, we show that the Sia system is important for producing low levels of c-di-GMP when P. aeruginosa is growing planktonically. In addition, we show that Sia activity is important for maintaining cell-associated Psl in planktonic populations. We also demonstrate that Sia mutant strains have reduced cell-associated Psl and a surface attachment-deficient phenotype. The Sia system also appears to posttranslationally impact cell-associated Psl levels. Collectively, our findings suggest a novel role for the Sia system and c-di-GMP in planktonic populations by regulating levels of cell-associated Psl.
Keywords: Pseudomonas aeruginosa; biofilm formation; c-di-GMP.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

