Identifying the Gene Regulatory Network of the Starvation-Induced Transcriptional Activator Nla28
- PMID: 36448789
- PMCID: PMC9765219
- DOI: 10.1128/jb.00265-22
Identifying the Gene Regulatory Network of the Starvation-Induced Transcriptional Activator Nla28
Abstract
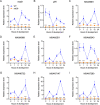
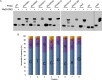
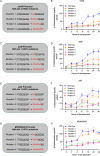
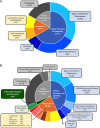
Myxococcus xanthus copes with starvation by producing fruiting bodies filled with dormant and stress-resistant spores. Here, we aimed to better define the gene regulatory network associated with Nla28, a transcriptional activator/enhancer binding protein (EBP) and a key regulator of the early starvation response. Previous work showed that Nla28 directly regulates EBP genes that are important for fruiting body development. However, the Nla28 regulatory network is likely to be much larger because hundreds of starvation-induced genes are downregulated in a nla28 mutant strain. To identify candidates for direct Nla28-mediated transcription, we analyzed the downregulated genes using a bioinformatics approach. Nine potential Nla28 target promoters (29 genes) were discovered. The results of in vitro promoter binding assays, coupled with in vitro and in vivo mutational analyses, suggested that the nine promoters along with three previously identified EBP gene promoters were indeed in vivo targets of Nla28. These results also suggested that Nla28 used tandem, imperfect repeats of an 8-bp sequence for promoter binding. Interestingly, eight of the new Nla28 target promoters were predicted to be intragenic. Based on mutational analyses, the newly identified Nla28 target loci contained at least one gene that was important for starvation-induced development. Most of these loci contained genes predicted to be involved in metabolic or defense-related functions. Using the consensus Nla28 binding sequence, bioinformatics, and expression profiling, 58 additional promoters and 102 genes were tagged as potential Nla28 targets. Among these putative Nla28 targets, functions, such as regulatory, metabolic, and cell envelope biogenesis, were assigned to many genes. IMPORTANCE In bacteria, starvation leads to profound changes in behavior and physiology. Some of these changes have economic and health implications because the starvation response has been linked to the formation of biofilms, virulence, and antibiotic resistance. To better understand how starvation contributes to changes in bacterial physiology and resistance, we identified the putative starvation-induced gene regulatory network associated with Nla28, a transcriptional activator from the bacterium Myxoccocus xanthus. We determined the mechanism by which starvation-responsive genes were activated by Nla28 and showed that several of the genes were important for the formation of a highly resistant cell type.
Keywords: biofilms; enhancer-binding proteins; fruiting body development; transcriptional activators; σ54 promoters.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

