Different Mechanisms Are Utilized by Coronavirus Transmissible Gastroenteritis Virus To Regulate Interferon Lambda 1 and Interferon Lambda 3 Production
- PMID: 36448799
- PMCID: PMC9769389
- DOI: 10.1128/jvi.01388-22
Different Mechanisms Are Utilized by Coronavirus Transmissible Gastroenteritis Virus To Regulate Interferon Lambda 1 and Interferon Lambda 3 Production
Abstract
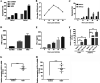
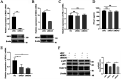
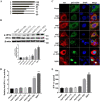
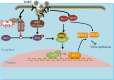
Type III interferons (IFN-λ) are shown to be preferentially produced by epithelial cells, which provide front-line protection at barrier surfaces. Transmissible gastroenteritis virus (TGEV), belonging to the genus Alphacoronavirus of the family Coronaviridae, can cause severe intestinal injuries in porcine, resulting in enormous economic losses for the swine industry, worldwide. Here, we demonstrated that although IFN-λ1 had a higher basal expression, TGEV infection induced more intense IFN-λ3 production in vitro and in vivo than did IFN-λ1. We explored the underlying mechanism of IFN-λ induction by TGEV and found a distinct regulation mechanism of IFN-λ1 and IFN-λ3. The classical RIG-I-like receptor (RLR) pathway is involved in IFN-λ3 but not IFN-λ1 production. Except for the signaling pathways mediated by RIG-I and MDA5, TGEV nsp1 induces IFN-λ1 and IFN-λ3 by activating NF-κB via the unfolded protein responses (UPR) PERK-eIF2α pathway. Furthermore, functional domain analysis indicated that the induction of IFN-λ by the TGEV nsp1 protein was located at amino acids 85 to 102 and was dependent on the phosphorylation of eIF2α and the nuclear translocation of NF-κB. Moreover, the recombinant TGEV with the altered amino acid motif of nsp1 85-102 was constructed, and the nsp1 (85-102sg) mutant virus significantly reduced the production of IFN-λ, compared with the wild strain. Compared to the antiviral activities of IFN-λ1, the administration of IFN-λ3 showed greater antiviral activity against TGEV infections in IPEC-J2 cells. In summary, our data point to the significant role of IFN-λ in the host innate antiviral responses to coronavirus infections within mucosal organs and in the distinct mechanisms of IFN-λ1 and IFN-λ3 regulation. IMPORTANCE Coronaviruses cause infectious diseases in various mammals and birds and exhibit an epithelial cell tropism in enteric and respiratory tracts. It is critical to explore how coronavirus infections modulate IFN-λ, a key innate cytokine against mucosal viral infection. Our results uncovered the different processes of IFN-λ1 and IFN-λ3 production that are involved in the classical RLR pathway and determined that TGEV nsp1 induces IFN-λ1 and IFN-λ3 production by activating NF-κB via the PERK-eIF2α pathway in UPR. These studies highlight the unique regulation of antiviral defense in the intestine during TGEV infection. We also demonstrated that IFN-λ3 induced greater antiviral activity against TGEV replication than did IFN-λ1 in IPEC-J2 cells, which is helpful in finding a novel strategy for the treatment of coronavirus infections.
Keywords: NF-κB; RLR-mediated signaling; coronavirus; nonstructural protein 1 (nsp1); protein kinase R-like ER kinase; transmissible gastroenteritis virus; type III IFN.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
The PERK Arm of the Unfolded Protein Response Negatively Regulates Transmissible Gastroenteritis Virus Replication by Suppressing Protein Translation and Promoting Type I Interferon Production.J Virol. 2018 Jul 17;92(15):e00431-18. doi: 10.1128/JVI.00431-18. Print 2018 Aug 1. J Virol. 2018. PMID: 29769338 Free PMC article.
-
Surface-Displayed Porcine IFN-λ3 in Lactobacillus plantarum Inhibits Porcine Enteric Coronavirus Infection of Porcine Intestinal Epithelial Cells.J Microbiol Biotechnol. 2020 Apr 28;30(4):515-525. doi: 10.4014/jmb.1909.09041. J Microbiol Biotechnol. 2020. PMID: 31838830 Free PMC article.
-
Porcine transmissible gastroenteritis virus nonstructural protein 2 contributes to inflammation via NF-κB activation.Virulence. 2018;9(1):1685-1698. doi: 10.1080/21505594.2018.1536632. Virulence. 2018. PMID: 30322331 Free PMC article.
-
Immune evasion of porcine enteric coronaviruses and viral modulation of antiviral innate signaling.Virus Res. 2016 Dec 2;226:128-141. doi: 10.1016/j.virusres.2016.05.015. Epub 2016 May 19. Virus Res. 2016. PMID: 27212682 Free PMC article. Review.
-
Porcine innate and adaptative immune responses to influenza and coronavirus infections.Ann N Y Acad Sci. 2006 Oct;1081(1):130-6. doi: 10.1196/annals.1373.014. Ann N Y Acad Sci. 2006. PMID: 17135502 Free PMC article. Review.
Cited by
-
Integrating network pharmacology with pharmacological research to elucidate the mechanism of modified Gegen Qinlian Decoction in treating porcine epidemic diarrhea.Sci Rep. 2024 Aug 15;14(1):18929. doi: 10.1038/s41598-024-70059-5. Sci Rep. 2024. PMID: 39147857 Free PMC article.
-
Transmissible gastroenteritis virus induces inflammatory responses via RIG-I/NF-κB/HIF-1α/glycolysis axis in intestinal organoids and in vivo.J Virol. 2024 Jun 13;98(6):e0046124. doi: 10.1128/jvi.00461-24. Epub 2024 May 23. J Virol. 2024. PMID: 38780247 Free PMC article.
-
MDA5 protein mediating persistent ER stress/unfolded protein response contributes to endothelial-mesenchymal-transition of lung microvascular endothelial cell in dermatomyositis.Cell Commun Signal. 2025 Mar 23;23(1):149. doi: 10.1186/s12964-025-02159-2. Cell Commun Signal. 2025. PMID: 40122798 Free PMC article.
References
-
- Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, Sorgeloos F, Ehl S, Mayer D, Kochs G, Schwemmle M, Gunther S, Drosten C, Michiels T, Staeheli P. 2010. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol 84:5670–5677. 10.1128/JVI.00272-10. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

