The SARS-CoV-2 spike protein binds and modulates estrogen receptors
- PMID: 36449624
- PMCID: PMC9710872
- DOI: 10.1126/sciadv.add4150
The SARS-CoV-2 spike protein binds and modulates estrogen receptors
Abstract
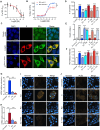
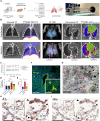
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) protein binds angiotensin-converting enzyme 2 as its primary infection mechanism. Interactions between S and endogenous proteins occur after infection but are not well understood. We profiled binding of S against >9000 human proteins and found an interaction between S and human estrogen receptor α (ERα). Using bioinformatics, supercomputing, and experimental assays, we identified a highly conserved and functional nuclear receptor coregulator (NRC) LXD-like motif on the S2 subunit. In cultured cells, S DNA transfection increased ERα cytoplasmic accumulation, and S treatment induced ER-dependent biological effects. Non-invasive imaging in SARS-CoV-2-infected hamsters localized lung pathology with increased ERα lung levels. Postmortem lung experiments from infected hamsters and humans confirmed an increase in cytoplasmic ERα and its colocalization with S in alveolar macrophages. These findings describe the discovery of a S-ERα interaction, imply a role for S as an NRC, and advance knowledge of SARS-CoV-2 biology and coronavirus disease 2019 pathology.
Figures


Update of
-
The SARS-CoV-2 spike protein binds and modulates estrogen receptors.bioRxiv [Preprint]. 2022 May 23:2022.05.21.492920. doi: 10.1101/2022.05.21.492920. bioRxiv. 2022. Update in: Sci Adv. 2022 Dec 2;8(48):eadd4150. doi: 10.1126/sciadv.add4150. PMID: 35665018 Free PMC article. Updated. Preprint.
References
-
- Mitrovic M., Sabljic N., Cvetkovic Z., Pantic N., Zivkovic Dakic A., Bukumiric Z., Libek V., Savic N., Milenkovic B., Virijevic M., Vucinic V., Milosevic I., Pravdic Z., Suvajdzic N., Fareed J., Antic D., Rotational thromboelastometry (ROTEM) profiling of COVID-19 patients. Platelets 32, 690–696 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

