Mitochondrial genome maintenance-the kinetoplast story
- PMID: 36449697
- PMCID: PMC10719067
- DOI: 10.1093/femsre/fuac047
Mitochondrial genome maintenance-the kinetoplast story
Abstract
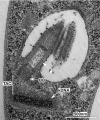
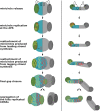
Mitochondrial DNA replication is an essential process in most eukaryotes. Similar to the diversity in mitochondrial genome size and organization in the different eukaryotic supergroups, there is considerable diversity in the replication process of the mitochondrial DNA. In this review, we summarize the current knowledge of mitochondrial DNA replication and the associated factors in trypanosomes with a focus on Trypanosoma brucei, and provide a new model of minicircle replication for this protozoan parasite. The model assumes the mitochondrial DNA (kinetoplast DNA, kDNA) of T. brucei to be loosely diploid in nature and the replication of the genome to occur at two replication centers at the opposing ends of the kDNA disc (also known as antipodal sites, APS). The new model is consistent with the localization of most replication factors and in contrast to the current model, it does not require the assumption of an unknown sorting and transport complex moving freshly replicated DNA to the APS. In combination with the previously proposed sexual stages of the parasite in the insect vector, the new model provides a mechanism for maintenance of the mitochondrial genetic diversity.
Keywords: kDNA; kinetoplast; mitochondrial DNA; mitochondrial DNA replication; trypanosomes.
© The Author(s) 2022. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Abu-Elneel K, Kapeller I, Shlomai J. Universal minicircle sequence-binding protein, a sequence-specific DNA-binding protein that recognizes the two replication origins of the kinetoplast DNA minicircle. J Biol Chem. 1999;274:13419–26. - PubMed

