Longitudinal analysis of serum neutralization of SARS-CoV-2 Omicron BA.2, BA.4, and BA.5 in patients receiving monoclonal antibodies
- PMID: 36450283
- PMCID: PMC9706550
- DOI: 10.1016/j.xcrm.2022.100850
Longitudinal analysis of serum neutralization of SARS-CoV-2 Omicron BA.2, BA.4, and BA.5 in patients receiving monoclonal antibodies
Abstract
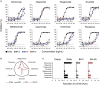
The emergence of Omicron sublineages impacts the therapeutic efficacy of anti-severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) monoclonal antibodies (mAbs). Here, we evaluate neutralization and antibody-dependent cellular cytotoxicity (ADCC) activities of 6 therapeutic mAbs against Delta, BA.2, BA.4, and BA.5. The Omicron subvariants escape most antibodies but remain sensitive to bebtelovimab and cilgavimab. Consistent with their shared spike sequence, BA.4 and BA.5 display identical neutralization profiles. Sotrovimab is the most efficient at eliciting ADCC. We also analyze 121 sera from 40 immunocompromised individuals up to 6 months after infusion of Ronapreve (imdevimab + casirivimab) or Evusheld (cilgavimab + tixagevimab). Sera from Ronapreve-treated individuals do not neutralize Omicron subvariants. Evusheld-treated individuals neutralize BA.2 and BA.5, but titers are reduced. A longitudinal evaluation of sera from Evusheld-treated patients reveals a slow decay of mAb levels and neutralization, which is faster against BA.5. Our data shed light on antiviral activities of therapeutic mAbs and the duration of effectiveness of Evusheld pre-exposure prophylaxis.
Keywords: ADCC; Omicron; SARS-CoV-2; antibodies; neutralization.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests T.B., C.P., H.M., and O.S. have a pending patent application for an anti-RBD mAb not used in this study (PCT/FR2021/070522).
Figures


References
-
- VanBlargan L.A., Errico J.M., Halfmann P.J., Zost S.J., Crowe J.E., Purcell L.A., Kawaoka Y., Corti D., Fremont D.H., Diamond M.S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 2022;28:490–495. doi: 10.1038/s41591-021-01678-y. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

