Undecaprenyl phosphate translocases confer conditional microbial fitness
- PMID: 36450355
- PMCID: PMC9876793
- DOI: 10.1038/s41586-022-05569-1
Undecaprenyl phosphate translocases confer conditional microbial fitness
Abstract
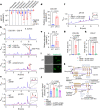
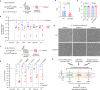
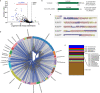
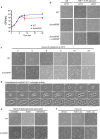
The microbial cell wall is essential for maintenance of cell shape and resistance to external stressors1. The primary structural component of the cell wall is peptidoglycan, a glycopolymer with peptide crosslinks located outside of the cell membrane1. Peptidoglycan biosynthesis and structure are responsive to shifting environmental conditions such as pH and salinity2-6, but the mechanisms underlying such adaptations are incompletely understood. Precursors of peptidoglycan and other cell surface glycopolymers are synthesized in the cytoplasm and then delivered across the cell membrane bound to the recyclable lipid carrier undecaprenyl phosphate7 (C55-P, also known as UndP). Here we identify the DUF368-containing and DedA transmembrane protein families as candidate C55-P translocases, filling a critical gap in knowledge of the proteins required for the biogenesis of microbial cell surface polymers. Gram-negative and Gram-positive bacteria lacking their cognate DUF368-containing protein exhibited alkaline-dependent cell wall and viability defects, along with increased cell surface C55-P levels. pH-dependent synthetic genetic interactions between DUF368-containing proteins and DedA family members suggest that C55-P transporter usage is dynamic and modulated by environmental inputs. C55-P transporter activity was required by the cholera pathogen for growth and cell shape maintenance in the intestine. We propose that conditional transporter reliance provides resilience in lipid carrier recycling, bolstering microbial fitness both inside and outside the host.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





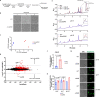


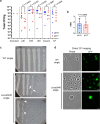
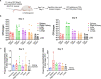
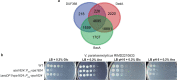
References
-
- Vijaranakul U, Nadakavukaren MJ, de Jonge BL, Wilkinson BJ, Jayaswal RK. Increased cell size and shortened peptidoglycan interpeptide bridge of NaCl-stressed Staphylococcus aureus and their reversal by glycine betaine. J. Bacteriol. 1995;177:5116–5121. doi: 10.1128/jb.177.17.5116-5121.1995. - DOI - PMC - PubMed
-
- Madiraju MVVS, Brunner DP, Wilkinson BJ. Effects of temperature, NaCl, and methicillin on penicillin-binding proteins, growth, peptidoglycan synthesis, and autolysis in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 1987;31:1727–1733. doi: 10.1128/AAC.31.11.1727. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

