Revisiting degron motifs in human AURKA required for its targeting by APC/CFZR1
- PMID: 36450448
- PMCID: PMC9713472
- DOI: 10.26508/lsa.202201372
Revisiting degron motifs in human AURKA required for its targeting by APC/CFZR1
Abstract
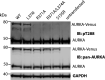
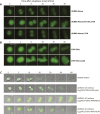
Mitotic kinase Aurora A (AURKA) diverges from other kinases in its multiple active conformations that may explain its interphase roles and the limited efficacy of drugs targeting the kinase pocket. Regulation of AURKA activity by the cell is critically dependent on destruction mediated by the anaphase-promoting complex (APC/CFZR1) during mitotic exit and G1 phase and requires an atypical N-terminal degron in AURKA called the "A-box" in addition to a reported canonical D-box degron in the C-terminus. Here, we find that the reported C-terminal D-box of AURKA does not act as a degron and instead mediates essential structural features of the protein. In living cells, the N-terminal intrinsically disordered region of AURKA containing the A-box is sufficient to confer FZR1-dependent mitotic degradation. Both in silico and in cellulo assays predict the QRVL short linear interacting motif of the A-box to be a phospho-regulated D-box. We propose that degradation of full-length AURKA also depends on an intact C-terminal domain because of critical conformational parameters permissive for both activity and mitotic degradation of AURKA.
© 2022 Abdelbaki et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures








References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
