The E3 ubiquitin ligase WWP2 regulates pro-fibrogenic monocyte infiltration and activity in heart fibrosis
- PMID: 36450710
- PMCID: PMC9712659
- DOI: 10.1038/s41467-022-34971-6
The E3 ubiquitin ligase WWP2 regulates pro-fibrogenic monocyte infiltration and activity in heart fibrosis
Abstract
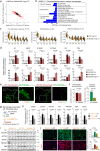
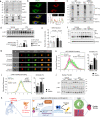
Non-ischemic cardiomyopathy (NICM) can cause left ventricular dysfunction through interstitial fibrosis, which corresponds to the failure of cardiac tissue remodeling. Recent evidence implicates monocytes/macrophages in the etiopathology of cardiac fibrosis, but giving their heterogeneity and the antagonizing roles of macrophage subtypes in fibrosis, targeting these cells has been challenging. Here we focus on WWP2, an E3 ubiquitin ligase that acts as a positive genetic regulator of human and murine cardiac fibrosis, and show that myeloid specific deletion of WWP2 reduces cardiac fibrosis in hypertension-induced NICM. By using single cell RNA sequencing analysis of immune cells in the same model, we establish the functional heterogeneity of macrophages and define an early pro-fibrogenic phase of NICM that is driven by Ccl5-expressing Ly6chigh monocytes. Among cardiac macrophage subtypes, WWP2 dysfunction primarily affects Ly6chigh monocytes via modulating Ccl5, and consequentially macrophage infiltration and activation, which contributes to reduced myofibroblast trans-differentiation. WWP2 interacts with transcription factor IRF7, promoting its non-degradative mono-ubiquitination, nuclear translocation and transcriptional activity, leading to upregulation of Ccl5 at transcriptional level. We identify a pro-fibrogenic macrophage subtype in non-ischemic cardiomyopathy, and demonstrate that WWP2 is a key regulator of IRF7-mediated Ccl5/Ly6chigh monocyte axis in heart fibrosis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

