A nuclease-mimetic platinum nanozyme induces concurrent DNA platination and oxidative cleavage to overcome cancer drug resistance
- PMID: 36450764
- PMCID: PMC9712435
- DOI: 10.1038/s41467-022-35022-w
A nuclease-mimetic platinum nanozyme induces concurrent DNA platination and oxidative cleavage to overcome cancer drug resistance
Abstract
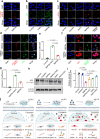
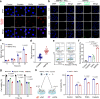
Platinum (Pt) resistance in cancer almost inevitably occurs during clinical Pt-based chemotherapy. The spontaneous nucleotide-excision repair of cancer cells is a representative process that leads to Pt resistance, which involves the local DNA bending to facilitate the recruitment of nucleotide-excision repair proteins and subsequent elimination of Pt-DNA adducts. By exploiting the structural vulnerability of this process, we herein report a nuclease-mimetic Pt nanozyme that can target cancer cell nuclei and induce concurrent DNA platination and oxidative cleavage to overcome Pt drug resistance. We show that the Pt nanozyme, unlike cisplatin and conventional Pt nanoparticles, specifically induces the nanozyme-catalyzed cleavage of the formed Pt-DNA adducts by generating in situ reactive oxygen species, which impairs the damage recognition factors-induced DNA bending prerequisite for nucleotide-excision repair. The recruitment of downstream effectors of nucleotide-excision repair to DNA lesion sites, including xeroderma pigmentosum groups A and F, is disrupted by the Pt nanozyme in cisplatin-resistant cancer cells, allowing excessive accumulation of the Pt-DNA adducts for highly efficient cancer therapy. Our study highlights the potential benefits of applying enzymatic activities to the use of the Pt nanomedicines, providing a paradigm shift in DNA damaging chemotherapy.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Reversal of platinum drug resistance by the histone deacetylase inhibitor belinostat.Lung Cancer. 2017 Jan;103:58-65. doi: 10.1016/j.lungcan.2016.11.019. Epub 2016 Nov 28. Lung Cancer. 2017. PMID: 28024697
-
A Platinum(IV) Anticancer Prodrug Targeting Nucleotide Excision Repair To Overcome Cisplatin Resistance.Angew Chem Int Ed Engl. 2016 Dec 12;55(50):15564-15568. doi: 10.1002/anie.201608936. Epub 2016 Oct 13. Angew Chem Int Ed Engl. 2016. PMID: 27736029
-
Polymerization by DNA polymerase eta is blocked by cis-diamminedichloroplatinum(II) 1,3-d(GpTpG) cross-link: implications for cytotoxic effects in nucleotide excision repair-negative tumor cells.Carcinogenesis. 2010 Mar;31(3):388-93. doi: 10.1093/carcin/bgp316. Epub 2009 Dec 16. Carcinogenesis. 2010. PMID: 20015866
-
Platinum-DNA adduct, nucleotide excision repair and platinum based anti-cancer chemotherapy.Cancer Treat Rev. 1998 Oct;24(5):331-44. doi: 10.1016/s0305-7372(98)90056-1. Cancer Treat Rev. 1998. PMID: 9861196 Review.
-
Genetic testing for chemotherapy in non-small cell lung cancer.Lung Cancer. 2003 Aug;41 Suppl 1:S97-102. doi: 10.1016/s0169-5002(03)00151-x. Lung Cancer. 2003. PMID: 12867068 Review.
Cited by
-
Metal-ligand cross-link strategy engineered iron-doped dopamine-based superstructure as peroxidase-like nanozymes for detection of glucose.Anal Bioanal Chem. 2024 Nov;416(27):6125-6136. doi: 10.1007/s00216-024-05317-6. Epub 2024 May 13. Anal Bioanal Chem. 2024. PMID: 38739158
-
Corosolic acid increases the therapeutic effect of cisplatin on gastric cancer by regulating Gpx4-dependent ferroptosis.Cancer Drug Resist. 2025 Aug 7;8:40. doi: 10.20517/cdr.2025.94. eCollection 2025. Cancer Drug Resist. 2025. PMID: 40843357 Free PMC article.
-
A Rational Design of Metal-Organic Framework Nanozyme with High-Performance Copper Active Centers for Alleviating Chemical Corneal Burns.Nanomicro Lett. 2023 Apr 30;15(1):112. doi: 10.1007/s40820-023-01059-9. Nanomicro Lett. 2023. PMID: 37121915 Free PMC article.
-
Self-propelled assembly of nanoparticles with self-catalytic regulation for tumour-specific imaging and therapy.Nat Commun. 2024 Jan 11;15(1):460. doi: 10.1038/s41467-024-44736-y. Nat Commun. 2024. PMID: 38212655 Free PMC article.
-
Skp2-mediated MLKL degradation confers cisplatin-resistant in non-small cell lung cancer cells.Commun Biol. 2023 Aug 2;6(1):805. doi: 10.1038/s42003-023-05166-6. Commun Biol. 2023. PMID: 37532777 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

