Deciphering the heterogeneity of the Lyve1+ perivascular macrophages in the mouse brain
- PMID: 36450771
- PMCID: PMC9712536
- DOI: 10.1038/s41467-022-35166-9
Deciphering the heterogeneity of the Lyve1+ perivascular macrophages in the mouse brain
Abstract
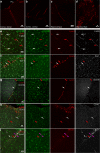
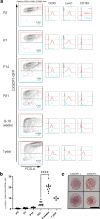
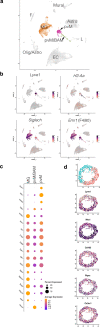
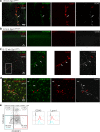
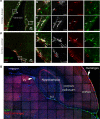
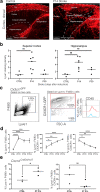
Perivascular macrophages (pvMs) are associated with cerebral vasculature and mediate brain drainage and immune regulation. Here, using reporter mouse models, whole brain and section immunofluorescence, flow cytometry, and single cell RNA sequencing, besides the Lyve1+F4/80+CD206+CX3CR1+ pvMs, we identify a CX3CR1- pvM population that shares phagocytic functions and location. Furthermore, the brain parenchyma vasculature mostly hosts Lyve1+MHCII- pvMs with low to intermediate CD45 expression. Using the double Cx3cr1GFP x Cx3cr1-Cre;RosatdT reporter mice for finer mapping of the lineages, we establish that CD45lowCX3CR1- pvMs are derived from CX3CR1+ precursors and require PU.1 during their ontogeny. In parallel, results from the Cxcr4-CreErt2;Rosa26tdT lineage tracing model support a bone marrow-independent replenishment of all Lyve1+ pvMs in the adult mouse brain. Lastly, flow cytometry and 3D immunofluorescence analysis uncover increased percentage of pvMs following photothrombotic induced stroke. Our results thus show that the parenchymal pvM population is more heterogenous than previously described, and includes a CD45low and CX3CR1- pvM population.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Ahn, J. H. et al. Meningeal lymphatic vessels at the skull base drain cerebrospinal fluid. Nature572, 62–66 (2019). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

