A Shigella sonnei clone with extensive drug resistance associated with waterborne outbreaks in China
- PMID: 36450777
- PMCID: PMC9709761
- DOI: 10.1038/s41467-022-35136-1
A Shigella sonnei clone with extensive drug resistance associated with waterborne outbreaks in China
Abstract
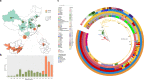
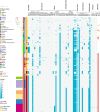
Antimicrobial resistance of Shigella sonnei has become a global concern. Here, we report a phylogenetic group of S. sonnei with extensive drug resistance, including a combination of multidrug resistance, coresistance to ceftriaxone and azithromycin (cefRaziR), reduced susceptibility to fluoroquinolones, and even colistin resistance (colR). This distinct clone caused six waterborne shigellosis outbreaks in China from 2015 to 2020. We collect 155 outbreak isolates and 152 sporadic isolates. The cefRaziR isolates, including outbreak strains, are mainly distributed in a distinct clade located in global Lineage III. The outbreak strains form a recently derived monophyletic group that may have emerged circa 2010. The cefRaziR and colR phenotypes are attributed to the acquisition of different plasmids, particularly the IncB/O/K/Z plasmid coharboring the blaCTX-M-14, mphA, aac(3)-IId, dfrA17, aadA5, and sul1 genes and the IncI2 plasmid with an mcr-1 gene. Genetic analyses identify 92 accessory genes and 60 single-nucleotide polymorphisms associated with the cefRaziR phenotype. Surveillance of this clone is required to determine its dissemination and threat to global public health.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

