A second look at exome sequencing data: detecting mobile elements insertion in a rare disease cohort
- PMID: 36450799
- PMCID: PMC10326243
- DOI: 10.1038/s41431-022-01250-3
A second look at exome sequencing data: detecting mobile elements insertion in a rare disease cohort
Abstract
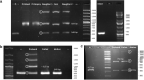
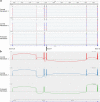
About 0.3% of all variants are due to de novo mobile element insertions (MEIs). The massive development of next-generation sequencing has made it possible to identify MEIs on a large scale. We analyzed exome sequencing (ES) data from 3232 individuals (2410 probands) with developmental and/or neurological abnormalities, with MELT, a tool designed to identify MEIs. The results were filtered by frequency, impacted region and gene function. Following phenotype comparison, two candidates were identified in two unrelated probands. The first mobile element (ME) was found in a patient referred for poikilodermia. A homozygous insertion was identified in the FERMT1 gene involved in Kindler syndrome. RNA study confirmed its pathological impact on splicing. The second ME was a de novo Alu insertion in the GRIN2B gene involved in intellectual disability, and detected in a patient with a developmental disorder. The frequency of de novo exonic MEIs in our study is concordant with previous studies on ES data. This project, which aimed to identify pathological MEIs in the coding sequence of genes, confirms that including detection of MEs in the ES pipeline can increase the diagnostic rate. This work provides additional evidence that ES could be used alone as a diagnostic exam.
© 2022. The Author(s), under exclusive licence to European Society of Human Genetics.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

