Cell type diversity in a developing octopus brain
- PMID: 36450803
- PMCID: PMC9712504
- DOI: 10.1038/s41467-022-35198-1
Cell type diversity in a developing octopus brain
Abstract
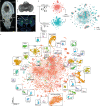
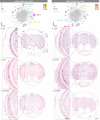
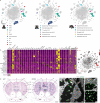
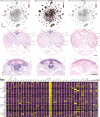
Octopuses are mollusks that have evolved intricate neural systems comparable with vertebrates in terms of cell number, complexity and size. The brain cell types that control their sophisticated behavioral repertoire are still unknown. Here, we profile the cell diversity of the paralarval Octopus vulgaris brain to build a cell type atlas that comprises mostly neural cells, but also multiple glial subtypes, endothelial cells and fibroblasts. We spatially map cell types to the vertical, subesophageal and optic lobes. Investigation of cell type conservation reveals a shared gene signature between glial cells of mouse, fly and octopus. Genes related to learning and memory are enriched in vertical lobe cells, which show molecular similarities with Kenyon cells in Drosophila. We construct a cell type taxonomy revealing transcriptionally related cell types, which tend to appear in the same brain region. Together, our data sheds light on cell type diversity and evolution in the octopus brain.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

