Common and rare variant associations with clonal haematopoiesis phenotypes
- PMID: 36450978
- PMCID: PMC9713173
- DOI: 10.1038/s41586-022-05448-9
Common and rare variant associations with clonal haematopoiesis phenotypes
Erratum in
-
Author Correction: Common and rare variant associations with clonal haematopoiesis phenotypes.Nature. 2023 Mar;615(7950):E3. doi: 10.1038/s41586-023-05803-4. Nature. 2023. PMID: 36807635 Free PMC article. No abstract available.
-
Author Correction: Common and rare variant associations with clonal haematopoiesis phenotypes.Nature. 2025 Jan;637(8047):E27. doi: 10.1038/s41586-024-08572-w. Nature. 2025. PMID: 39779868 No abstract available.
Abstract
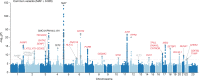
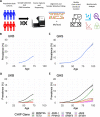
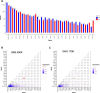
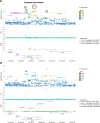
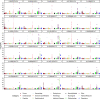
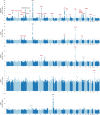
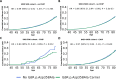
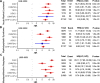
Clonal haematopoiesis involves the expansion of certain blood cell lineages and has been associated with ageing and adverse health outcomes1-5. Here we use exome sequence data on 628,388 individuals to identify 40,208 carriers of clonal haematopoiesis of indeterminate potential (CHIP). Using genome-wide and exome-wide association analyses, we identify 24 loci (21 of which are novel) where germline genetic variation influences predisposition to CHIP, including missense variants in the lymphocytic antigen coding gene LY75, which are associated with reduced incidence of CHIP. We also identify novel rare variant associations with clonal haematopoiesis and telomere length. Analysis of 5,041 health traits from the UK Biobank (UKB) found relationships between CHIP and severe COVID-19 outcomes, cardiovascular disease, haematologic traits, malignancy, smoking, obesity, infection and all-cause mortality. Longitudinal and Mendelian randomization analyses revealed that CHIP is associated with solid cancers, including non-melanoma skin cancer and lung cancer, and that CHIP linked to DNMT3A is associated with the subsequent development of myeloid but not lymphoid leukaemias. Additionally, contrary to previous findings from the initial 50,000 UKB exomes6, our results in the full sample do not support a role for IL-6 inhibition in reducing the risk of cardiovascular disease among CHIP carriers. Our findings demonstrate that CHIP represents a complex set of heterogeneous phenotypes with shared and unique germline genetic causes and varied clinical implications.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
M.D.K., A.D., S.O., N.B., D.L., K.W., A.M., M.V.M., S.S., J.H., J.T., J.A.K., V.M.R., Y.Z., Y.H., A.G., C.G., J. Mbatchou, R.R.W., N.V., J.B., N.N.P., M.G.L., M.J., D.J.G., L.A.L., M.N.C., G.S.A., A.E.L., M.A.R.F., R.D., C.P., A.R.S., G.T., A.A.F., W.S., J.G.R., J.D.O., J. Marchini, H.M.K., A.B., G.R.A. and E.J. are current employees and/or stockholders of Regeneron Genetics Center or Regeneron Pharmaceuticals.
Figures




Comment in
-
CHIPping away at the genetic aetiology of clonal haematopoiesis.Nat Rev Genet. 2023 Feb;24(2):69. doi: 10.1038/s41576-022-00565-7. Nat Rev Genet. 2023. PMID: 36509999 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

