Gene expression profile for different susceptibilities to sound stimulation: a comparative study on brainstems between two inbred laboratory mouse strains
- PMID: 36451107
- PMCID: PMC9710100
- DOI: 10.1186/s12864-022-09016-3
Gene expression profile for different susceptibilities to sound stimulation: a comparative study on brainstems between two inbred laboratory mouse strains
Abstract
Background: DBA/1 mice have a higher susceptibility to generalized audiogenic seizures (AGSz) and seizure-induced respiratory arrest (S-IRA) than C57/BL6 mice. The gene expression profile might be potentially related to this difference. This study aimed to investigate the susceptibility difference in AGSz and S-IRA between DBA/1 and C57BL/6 mice by profiling long noncoding RNAs (lncRNAs) and mRNA expression.
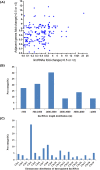
Methods: We compared lncRNAs and mRNAs from the brainstem of the two strains with Arraystar Mouse lncRNA Microarray V3.0 (Arraystar, Rockville, MD). Gene Ontology (GO) and pathway analyses were performed to determine the potentially related biological functions and pathways based on differentially expressed mRNAs. qRT-PCR was carried out to validate the results.
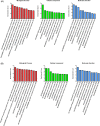
Results: A total of 897 lncRNAs and 438 mRNAs were differentially expressed (fold change ≥2, P < 0.05), of which 192 lncRNAs were upregulated and 705 lncRNAs were downregulated. A total of 138 mRNAs were upregulated, and 300 mRNAs were downregulated. In terms of specific mRNAs, Htr5b, Gabra2, Hspa1b and Gfra1 may be related to AGSz or S-IRA. Additionally, lncRNA Neat1 may participate in the difference in susceptibility. GO and pathway analyses suggested that TGF-β signaling, metabolic process and MHC protein complex could be involved in these differences. Coexpression analysis identified 9 differentially expressed antisense lncRNAs and 115 long intergenic noncoding RNAs (lincRNAs), and 2010012P19Rik and its adjacent RNA Tnfsf12-Tnfsf13 may have participated in S-IRA by regulating sympathetic neuron function. The results of the qRT-PCR of five selected lncRNAs (AK038711, Gm11762, 1500004A13Rik, AA388235 and Neat1) and four selected mRNAs (Hspa1b, Htr5b, Gabra2 and Gfra1) were consistent with those obtained by microarray.
Conclusion: We concluded that TGF-β signaling and metabolic process may contribute to the differential sensitivity to AGSz and S-IRA. Among mRNAs, Htr5b, Gabra2, Hspa1b and Gfra1 could potentially influence the susceptibility. LncRNA Neat1 and 2010012P19Rik may also contribute to the different response to sound stimulation. Further studies should be carried out to explore the underlying functions and mechanisms of differentially expressed RNAs.
Keywords: AGSz; Long noncoding RNAs; S-IRA; SUDEP model.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

