Modeling the impact of data sharing on variant classification
- PMID: 36451272
- PMCID: PMC9933054
- DOI: 10.1093/jamia/ocac232
Modeling the impact of data sharing on variant classification
Abstract
Objective: Many genetic variants are classified, but many more are variants of uncertain significance (VUS). Clinical observations of patients and their families may provide sufficient evidence to classify VUS. Understanding how long it takes to accumulate sufficient patient data to classify VUS can inform decisions in data sharing, disease management, and functional assay development.
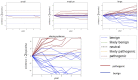
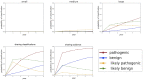
Materials and methods: Our software models the accumulation of clinical evidence (and excludes all other types of evidence) to measure their unique impact on variant interpretation. We illustrate the time and probability for VUS classification when laboratories share evidence, when they silo evidence, and when they share only variant interpretations.
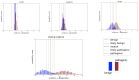
Results: Using conservative assumptions for frequencies of observed clinical evidence, our models show the probability of classifying rare pathogenic variants with an allele frequency of 1/100 000 increases from less than 25% with no data sharing to nearly 80% after one year when labs share data, with nearly 100% classification after 5 years. Conversely, our models found that extremely rare (1/1 000 000) variants have a low probability of classification using only clinical data.
Discussion: These results quantify the utility of data sharing and demonstrate the importance of alternative lines of evidence for interpreting rare variants. Understanding variant classification circumstances and timelines provides valuable insight for data owners, patients, and service providers. While our modeling parameters are based on our own assumptions of the rate of accumulation of clinical observations, users may download the software and run simulations with updated parameters.
Conclusions: The modeling software is available at https://github.com/BRCAChallenge/classification-timelines.
Keywords: benign; classification; genetic variation; modeling; pathogenic.
© The Author(s) 2022. Published by Oxford University Press on behalf of the American Medical Informatics Association. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures