Single-cell atlases link macrophages and CD8+ T-cell subpopulations to disease progression and immunotherapy response in urothelial carcinoma
- PMID: 36451860
- PMCID: PMC9706581
- DOI: 10.7150/thno.77281
Single-cell atlases link macrophages and CD8+ T-cell subpopulations to disease progression and immunotherapy response in urothelial carcinoma
Abstract
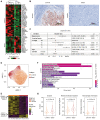
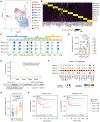
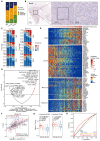
Rationale: Immune checkpoint inhibitors (ICIs) have revolutionized the management of locally advanced or metastatic urothelial carcinoma. Strikingly, compared to urothelial carcinoma of the bladder (UCB), upper tract urothelial carcinoma (UTUC) has a higher response rate to ICIs. The stratification of patients most likely to benefit from ICI therapy remains a major clinical challenge. Methods: In this study, we performed the first single-cell RNA sequencing (scRNA-seq) study of 13 surgical tissue specimens from 12 patients with UTUC. The key results were validated by the analysis of two independent cohorts with bulk RNA-seq data for UCB (n = 404) and UTUC (n = 158) and one cohort of patients with metastatic urothelial carcinoma (mUC) who were treated with atezolizumab (n = 348). Results: Using scRNA-seq, we observed a higher proportion of tumor-infiltrating immune cells in locally advanced UTUC. Similar prognostically relevant intrinsic basal and luminal-like epithelial subtypes were found in both UTUC and UCB, although UTUC is predominantly of the luminal subtype. We also discovered that immunosuppressive macrophages and exhausted T-cell subpopulations were enriched in the basal subtype and showed enhanced interactions. Furthermore, we developed a gene expression signature (Macro-C3 score) capturing the immunosuppressive macrophages that better predicts outcomes than the currently established subtypes. We also developed a computational method to model immune evasion, and the Macro-C3 score predicted therapeutic response of mUC treated with first-line anti-PD-L1 inhibitors in patients with lower basal scores. Conclusions: Overall, the distinct microenvironment and Macro-C3 score provide an explanation for ICI efficacy in urothelial carcinoma and reveal new candidate regulators of immune evasion, suggesting potential therapeutic targets for improving antitumor immunity in the basal subtype.
Keywords: Immunosuppression; Immunotherapy; Single-cell RNA sequencing; Tumor microenvironment; Urothelial carcinoma.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



References
-
- Roupret M, Babjuk M, Burger M, Capoun O, Cohen D, Comperat EM. et al. European Association of Urology Guidelines on Upper Urinary Tract Urothelial Carcinoma: 2020 Update. Eur Urol. 2021;79:62–79. - PubMed
-
- Margulis V, Shariat SF, Matin SF, Kamat AM, Zigeuner R, Kikuchi E. et al. Outcomes of radical nephroureterectomy: a series from the Upper Tract Urothelial Carcinoma Collaboration. Cancer. 2009;115:1224–33. - PubMed
-
- Moss TJ, Qi Y, Xi L, Peng B, Kim TB, Ezzedine NE. et al. Comprehensive Genomic Characterization of Upper Tract Urothelial Carcinoma. Eur Urol. 2017;72:641–9. - PubMed
-
- Necchi A, Madison R, Pal SK, Ross JS, Agarwal N, Sonpavde G. et al. Comprehensive Genomic Profiling of Upper-tract and Bladder Urothelial Carcinoma. Eur Urol Focus. 2021;7:1339–46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

