Janus USPION modular platform (JUMP) for theranostic ultrasound-mediated targeted intratumoral microvascular imaging and DNA/miRNA delivery
- PMID: 36451861
- PMCID: PMC9706579
- DOI: 10.7150/thno.78454
Janus USPION modular platform (JUMP) for theranostic ultrasound-mediated targeted intratumoral microvascular imaging and DNA/miRNA delivery
Abstract
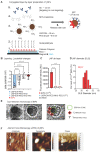
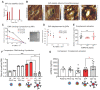
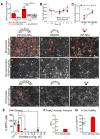
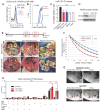
Rationale: High mortality in pancreatic cancer (PDAC) and triple negative breast cancer (TNBC) highlight the need to capitalize on nanoscale-design advantages for multifunctional diagnostics and therapies. DNA/RNA-therapies can provide potential breakthroughs, however, to date, there is no FDA-approved systemic delivery system to solid tumors. Methods: Here, we report a Janus-nanoparticle (jNP)-system with modular targeting, payload-delivery, and targeted-imaging capabilities. Our jNP-system consists of 10 nm ultrasmall superparamagnetic iron oxide nanoparticles (USPION) with opposing antibody-targeting and DNA/RNA payload-protecting faces, directionally self-assembled with commercially available zwitterionic microbubbles (MBs) and DNA/RNA payloads. Results: Sonoporation of targeted jNP-payload-MBs delivers functional reporter-DNA imparting tumor-fluorescence, and micro-RNA126 reducing non-druggable KRAS in PDAC-Panc1 and TNBC-MB231 xenografted tumors. The targeting jNP-system enhances ultrasound-imaging of intra-tumoral microvasculature using less MBs/body weight (BW). The jNP-design enhances USPION's T2*-magnetic resonance (MR) and MR-imaging of PDAC-peritoneal metastases using less Fe/BW. Conclusion: Altogether, data advance the asymmetric jNP-design as a potential theranostic Janus-USPION Modular Platform - a JUMP forward.
Keywords: Janus nanoparticle; Nanomedicine; USPION; modular nanotheranostics; nano-micro hybrid platform; nucleic acid delivery; pancreatic cancer.
© The author(s).
Conflict of interest statement
Competing Interests: Awarded patents pertaining to the results presented in the paper: J.Y. Wong, R.D. Whitaker, N. Ruiz-Opazo, and V.L.M. Herrera, PCT Application No. PCT/US2016/018417, Theranostic compositions and uses thereof, International Filing Date: Feb 18, 2016 - Assigned U.S. Patent No. 10,568,970, ISSUED February 25, 2020. Boston University holds awarded and pending patents on DEspR (dual endothelin1/VEGFsp receptor). VLMH and NRO are co-inventors on DEspR patents filed by Boston University. All other authors declare they have no competing interests.
Figures


References
-
- Li X, Yang J, Peng L, Sahin AA, Huo L, Ward KC. et al. Triple-negative breast cancer has worse overall survival and cause-specific survival than non-triple-negative breast cancer. Breast Cancer Res Treat. 2017;161:279–87. - PubMed
-
- Berrecoso G, Crecente-Campo J, Alonso MJ. Unveiling the pitfalls of the protein corona of polymeric drug nanocarriers. Drug Deliv Transl Res. 2020;10:730–50. - PubMed
-
- Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U. et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol. 2013;31:638–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

