Effects of trichothecene production by Trichoderma arundinaceum isolates from bean-field soils on the defense response, growth and development of bean plants (Phaseolus vulgaris)
- PMID: 36452093
- PMCID: PMC9702529
- DOI: 10.3389/fpls.2022.1005906
Effects of trichothecene production by Trichoderma arundinaceum isolates from bean-field soils on the defense response, growth and development of bean plants (Phaseolus vulgaris)
Abstract
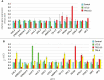
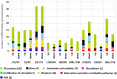
The trichothecene toxin-producing fungus Trichoderma arundinaceum has potential as a biological control agent. However, most biocontrol studies have focused only on one strain, IBT 40837. In the current study, three Trichoderma isolates recovered from bean-field soils produced the trichothecene harzianum A (HA) and trichodermol, the latter being an intermediate in the HA biosynthesis. Based on phylogenetic analysis, the three isolates were assigned to the species T. arundinaceum. Their genome sequences had a high degree of similarity to the reference IBT 40837 strain, in terms of total genome size, number of predicted genes, and diversity of putative secondary metabolite biosynthetic gene clusters. HA production by these bean-field isolates conferred significant in vitro antifungal activity against Rhizoctonia solani and Sclerotinia sclerotiorum, which are some of the most important bean pathogens. Furthermore, the bean-field isolates stimulated germination of bean seeds and subsequent growth of above ground parts of the bean plant. Transcriptomic analysis of bean plants inoculated with these T. arundinaceum bean-field soil isolates indicated that HA production significantly affected expression of plant defense-related genes; this effect was particularly significant in the expression of chitinase-encoding genes. Together, these results indicate that Trichoderma species producing non-phytotoxic trichothecenes can induce defenses in plants without negatively affecting germination and development.
Keywords: biological control; gene-clusters; metabolomics; plant-fungal interaction; secondary metabolites; sesquiterpenes; transcriptomics.
Copyright © 2022 Cardoza, Mayo-Prieto, Martínez-Reyes, McCormick, Carro-Huerga, Campelo, Rodríguez-González, Lorenzana, Proctor, Casquero and Gutiérrez.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures









References
-
- APS (2010). Compendium of bean diseases. (Minnesota, USA: The American Phytopathological Society; ).
-
- Becares A., Fernández A. F. (2017). Microbiome based identification, monitoring and enhancement of fermentation processes and products. International Patent WO 2017/096385 A1.
-
- Benitez T., Rincón A. M., Limón M. C., Codón A. C. (2004). Biocontrol mechanisms of Trichoderma strains. Int. Microbiol. 7 (4), 249–260. - PubMed
LinkOut - more resources
Full Text Sources

