Comparing adventitious root-formation and graft-unification abilities in clones of Argania spinosa
- PMID: 36452103
- PMCID: PMC9702570
- DOI: 10.3389/fpls.2022.1002703
Comparing adventitious root-formation and graft-unification abilities in clones of Argania spinosa
Abstract
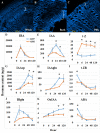
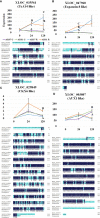
Argania spinosa trees have attracted attention in recent years due to their high resistance to extreme climate conditions. Initial domestication activities practiced in Morocco. Here we report on selection and vegetative propagation of A. spinosa trees grown in Israel. Trees yielding relatively high amounts of fruit were propagated by rooting of stem cuttings. High variability in rooting ability was found among the 30 clones selected. In-depth comparison of a difficult-to-root (ARS7) and easy-to-root (ARS1) clone revealed that the rooted cuttings of ARS7 have a lower survival rate than those of ARS1. In addition, histological analysis of the adventitious root primordia showed many abnormal fused primordia in ARS7. Hormone profiling revealed that while ARS1 accumulates more cytokinin, ARS7 accumulates more auxin, suggesting different auxin-to-cytokinin ratios underlying the different rooting capabilities. The hypothesized relationship between rooting and grafting abilities was addressed. Reciprocal grafting was performed with ARS1/ARS7 but no significant differences in the success of graft unification between the trees was detected. Accordingly, comparative RNA sequencing of the rooting and grafting zones showed more differentially expressed genes related to rooting than to grafting between the two trees. Clustering, KEGG and Venn analyses confirmed enrichment of genes related to auxin metabolism, transport and signaling, cytokinin metabolism and signaling, cell wall modification and cell division in both regions. In addition, the differential expression of some key genes in ARS1 vs. ARS7 rooting zones was revealed. Taken together, while both adventitious root-formation and graft-unification processes share response to wounding, cell reprogramming, cell division, cell differentiation and reconnection of the vasculature, there are similar, but also many different genes regulating the two processes. Therefore an individual genotype can have low rooting capacity but good graft-unification ability.
Keywords: adventitious roots; argan; grafting; hormone profile; transcription profile.
Copyright © 2022 Tzeela, Yechezkel, Serero, Eliyahu, Sherf, Manni, Doron-Faigenboim, Carmelli-Weissberg, Shaya, Dwivedi and Sadot.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Early steps of adventitious rooting: morphology, hormonal profiling and carbohydrate turnover in carnation stem cuttings.Physiol Plant. 2014 Mar;150(3):446-62. doi: 10.1111/ppl.12114. Epub 2013 Oct 24. Physiol Plant. 2014. PMID: 24117983
-
Temporal profiling of physiological, histological, and transcriptomic dissection during auxin-induced adventitious root formation in tetraploid Robinia pseudoacacia micro-cuttings.Planta. 2024 Feb 8;259(3):66. doi: 10.1007/s00425-024-04341-1. Planta. 2024. PMID: 38332379
-
Distinct effects of auxin and light on adventitious root development in Eucalyptus saligna and Eucalyptus globulus.Tree Physiol. 2001 May;21(7):457-64. doi: 10.1093/treephys/21.7.457. Tree Physiol. 2001. PMID: 11340046
-
Plant Hormone Homeostasis, Signaling, and Function during Adventitious Root Formation in Cuttings.Front Plant Sci. 2016 Mar 31;7:381. doi: 10.3389/fpls.2016.00381. eCollection 2016. Front Plant Sci. 2016. PMID: 27064322 Free PMC article. Review.
-
Adventitious Rooting in Populus Species: Update and Perspectives.Front Plant Sci. 2021 May 20;12:668837. doi: 10.3389/fpls.2021.668837. eCollection 2021. Front Plant Sci. 2021. PMID: 34093625 Free PMC article. Review.
Cited by
-
Slow release of a synthetic auxin induces formation of adventitious roots in recalcitrant woody plants.Nat Biotechnol. 2024 Nov;42(11):1705-1716. doi: 10.1038/s41587-023-02065-3. Epub 2024 Jan 24. Nat Biotechnol. 2024. PMID: 38267759
References
-
- Abu-Abied M., Szwerdszarf D., Mordehaev I., Levy A., Stelmakh O. R., Belausov E., et al. . (2012). Microarray analysis revealed upregulation of nitrate reductase in juvenile cuttings of eucalyptus grandis, which correlated with increased nitric oxide production and adventitious root formation. Plant J. 71, 787–799. doi: 10.1111/j.1365-313X.2012.05032.x - DOI - PubMed
-
- Ait Hammou R., Harrouni M. C., Cherifi K., Hallouti A., Daoud S. (2018). Clonal selection of argan trees (Argania spinosa l. skeels) in the admin forest, agadir, southwestern Morocco. J. Agric. Crop Res. 6 (1), 7–18. doi: 10.33495/jacr_v6i1.18.110 - DOI
LinkOut - more resources
Full Text Sources

