PP2A phosphatase regulates cell-type specific cytoskeletal organization to drive dendrite diversity
- PMID: 36452406
- PMCID: PMC9702092
- DOI: 10.3389/fnmol.2022.926567
PP2A phosphatase regulates cell-type specific cytoskeletal organization to drive dendrite diversity
Abstract
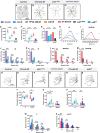
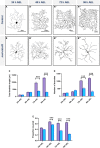
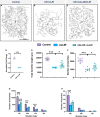
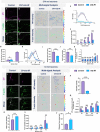
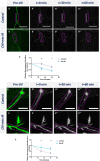
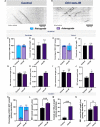
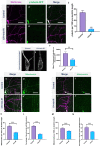
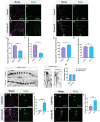
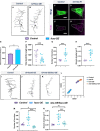
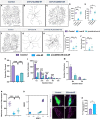
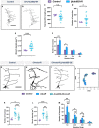
Uncovering molecular mechanisms regulating dendritic diversification is essential to understanding the formation and modulation of functional neural circuitry. Transcription factors play critical roles in promoting dendritic diversity and here, we identify PP2A phosphatase function as a downstream effector of Cut-mediated transcriptional regulation of dendrite development. Mutant analyses of the PP2A catalytic subunit (mts) or the scaffolding subunit (PP2A-29B) reveal cell-type specific regulatory effects with the PP2A complex required to promote dendritic growth and branching in Drosophila Class IV (CIV) multidendritic (md) neurons, whereas in Class I (CI) md neurons, PP2A functions in restricting dendritic arborization. Cytoskeletal analyses reveal requirements for Mts in regulating microtubule stability/polarity and F-actin organization/dynamics. In CIV neurons, mts knockdown leads to reductions in dendritic localization of organelles including mitochondria and satellite Golgi outposts, while CI neurons show increased Golgi outpost trafficking along the dendritic arbor. Further, mts mutant neurons exhibit defects in neuronal polarity/compartmentalization. Finally, genetic interaction analyses suggest β-tubulin subunit 85D is a common PP2A target in CI and CIV neurons, while FoxO is a putative target in CI neurons.
Keywords: Drosophila; cytoskeleton; dendrite; microtubules; neuronal polarity; protein phosphatase 2A; transcriptional regulation.
Copyright © 2022 Bhattacharjee, Lottes, Nanda, Golshir, Patel, Ascoli and Cox.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

