Computational approaches for network-based integrative multi-omics analysis
- PMID: 36452456
- PMCID: PMC9703081
- DOI: 10.3389/fmolb.2022.967205
Computational approaches for network-based integrative multi-omics analysis
Abstract
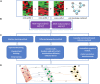
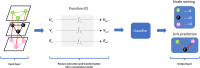
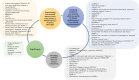
Advances in omics technologies allow for holistic studies into biological systems. These studies rely on integrative data analysis techniques to obtain a comprehensive view of the dynamics of cellular processes, and molecular mechanisms. Network-based integrative approaches have revolutionized multi-omics analysis by providing the framework to represent interactions between multiple different omics-layers in a graph, which may faithfully reflect the molecular wiring in a cell. Here we review network-based multi-omics/multi-modal integrative analytical approaches. We classify these approaches according to the type of omics data supported, the methods and/or algorithms implemented, their node and/or edge weighting components, and their ability to identify key nodes and subnetworks. We show how these approaches can be used to identify biomarkers, disease subtypes, crosstalk, causality, and molecular drivers of physiological and pathological mechanisms. We provide insight into the most appropriate methods and tools for research questions as showcased around the aetiology and treatment of COVID-19 that can be informed by multi-omics data integration. We conclude with an overview of challenges associated with multi-omics network-based analysis, such as reproducibility, heterogeneity, (biological) interpretability of the results, and we highlight some future directions for network-based integration.
Keywords: data integration; machine learning; multi-modal network; multi-omics; network causal inference; network diffusion/propagation.
Copyright © 2022 Agamah, Bayjanov, Niehues, Njoku, Skelton, Mazandu, Ederveen, Mulder, Chimusa and 't Hoen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources

