Inhibiting the biogenesis of myeloid-derived suppressor cells enhances immunotherapy efficacy against mammary tumor progression
- PMID: 36453551
- PMCID: PMC9711879
- DOI: 10.1172/JCI158661
Inhibiting the biogenesis of myeloid-derived suppressor cells enhances immunotherapy efficacy against mammary tumor progression
Abstract
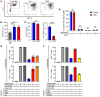
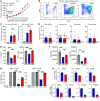
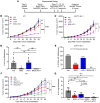
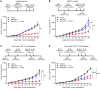
While immune checkpoint inhibitors (ICIs) have transformed the therapeutic landscape in oncology, they are effective in select subsets of patients. Efficacy may be limited by tumor-driven immune suppression, of which 1 key mechanism is the development of myeloid-derived suppressor cells (MDSCs). A fundamental gap in MDSC therapeutics is the lack of approaches that target MDSC biogenesis. We hypothesized that targeting MDSC biogenesis would mitigate MDSC burden and bolster tumor responses to ICIs. We tested a class of agents, dihydroorotate dehydrogenase (DHODH) inhibitors, that have been previously shown to restore the terminal differentiation of leukemic myeloid progenitors. DHODH inhibitors have demonstrated preclinical safety and are under clinical study for hematologic malignancies. Using mouse models of mammary cancer that elicit robust MDSC responses, we demonstrated that the DHODH inhibitor brequinar (a) suppressed MDSC production from early-stage myeloid progenitors, which was accompanied by enhanced myeloid maturation; (b) augmented the antitumor and antimetastatic activities of programmed cell death 1-based (PD-1-based) ICI therapy in ICI-resistant mammary cancer models; and (c) acted in concert with PD-1 blockade through modulation of MDSC and CD8+ T cell responses. Moreover, brequinar facilitated myeloid maturation and inhibited immune-suppressive features in human bone marrow culture systems. These findings advance the concept of MDSC differentiation therapy in immuno-oncology.
Keywords: Cancer immunotherapy; Immunology.
Figures


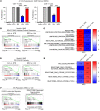
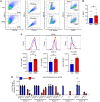
Comment in
- Bring on the brequinar: An approach to enforce differentiation of myeloid derived suppressor cells doi: 10.1172/JCI165506
References
-
- Grover A, et al. Myeloid-derived suppressor cells: a propitious road to clinic. Cancer Discov. 2021;11(11):2693–2706. doi: 10.1158/2159-8290.CD-21-0764. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

