Elevated RIPK3 correlates with disease burden in myelofibrosis
- PMID: 36453650
- PMCID: PMC10111350
- DOI: 10.1182/bloodadvances.2021006838
Elevated RIPK3 correlates with disease burden in myelofibrosis
Conflict of interest statement
Figures
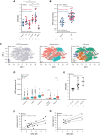
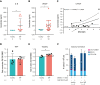
References
-
- Le Bousse-Kerdiles MC, Martyre MC. Dual implication of fibrogenic cytokines in the pathogenesis of fibrosis and myeloproliferation in myeloid metaplasia with myelofibrosis. Ann Hematol. 1999;78(10):437–444. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

