Pharokka: a fast scalable bacteriophage annotation tool
- PMID: 36453861
- PMCID: PMC9805569
- DOI: 10.1093/bioinformatics/btac776
Pharokka: a fast scalable bacteriophage annotation tool
Abstract
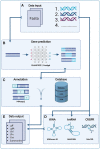
Summary: In recent years, there has been an increasing interest in bacteriophages, which has led to growing numbers of bacteriophage genomic sequences becoming available. Consequently, there is a need for a rapid and consistent genomic annotation tool dedicated for bacteriophages. Existing tools either are not designed specifically for bacteriophages or are web- and email-based and require significant manual curation, which makes their integration into bioinformatic pipelines challenging. Pharokka was created to provide a tool that annotates bacteriophage genomes easily, rapidly and consistently with standards compliant outputs. Moreover, Pharokka requires only two lines of code to install and use and takes under 5 min to run for an average 50-kb bacteriophage genome.
Availability and implementation: Pharokka is implemented in Python and is available as a bioconda package using 'conda install -c bioconda pharokka'. The source code is available on GitHub (https://github.com/gbouras13/pharokka). Pharokka has been tested on Linux-64 and MacOSX machines and on Windows using a Linux Virtual Machine.
© The Author(s) 2022. Published by Oxford University Press.
Figures