Direct Nasal Swab for Rapid Test and Saliva as an Alternative Biological Sample for RT-PCR in COVID-19 Diagnosis
- PMID: 36453913
- PMCID: PMC9769842
- DOI: 10.1128/spectrum.01998-22
Direct Nasal Swab for Rapid Test and Saliva as an Alternative Biological Sample for RT-PCR in COVID-19 Diagnosis
Erratum in
-
Erratum for Sazed et al., "Direct Nasal Swab for Rapid Test and Saliva as an Alternative Biological Sample for RT-PCR in COVID-19 Diagnosis".Microbiol Spectr. 2024 Mar 5;12(3):e0401423. doi: 10.1128/spectrum.04014-23. Epub 2024 Jan 22. Microbiol Spectr. 2024. PMID: 38251893 Free PMC article. No abstract available.
Abstract
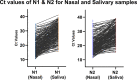
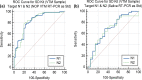
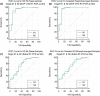
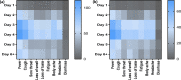
Accurate and early diagnoses are prerequisites for prompt treatment. For coronavirus disease 2019 (COVID-19), it is even more crucial. Currently, choice of methods include rapid diagnostic tests and reverse transcription polymerase chain reaction (RT-PCR) using samples mostly of respiratory origin and sometimes saliva. We evaluated two rapid diagnostic tests with three specimen types using viral transport medium (VTM) containing naso-oropharyngeal (NOP) swabs, direct nasal and direct nasopharyngeal (NP) samples from 428 prospective patients. We also performed RT-PCR for 428 NOP VTM and 316 saliva samples to compare results. The sensitivity of the SD Biosensor Standard Q COVID-19 antigen (Ag) test kit drastically raised from an average of 65.55% (NOP VTM) to 85.25% (direct nasal samples), while RT-PCR was the gold standard. For the CareStart kit, the sensitivity was almost similar for direct NP swabs; the average was 84.57%. The specificities were ≥95% for both SD Biosensor Standard Q and CareStart COVID-19 Ag tests in all platforms. The kits were also able to detect patients with different variants as well. Alternatively, RT-PCR results from saliva and NOP VTM samples showed high sensitivities of 96.45% and 95.48% with respect to each other as standard. The overall results demonstrated high performance of the rapid tests, indicating the suitability for regular surveillance at clinical facilities when using direct nasal or direct NP samples rather than NOP VTM. Additionally, the analysis also signifies not showed that RT-PCR of saliva can be used as an choice of method to RT-PCR of NOP VTM, providing an easier, non-invasive sample collection method. IMPORTANCE There are several methods for the diagnosis of coronavirus disease 2019 (COVID-19), and the choice of methods depends mostly on the resources and level of sensitivity required by the user and health care providers. Still, reverse transcription polymerase chain reaction (RT-PCR) has been chosen as the best method using direct naso-oropharyngeal swabs. There are also other methods of fast detection, such as rapid diagnostic tests (RDTs), which offer result within 15 to 20 min and have become quite popular for self-testing and in the clinical setting. The major drawback of the currently used RT-PCR method is compliance, as it may cause irritation, and patients often refuse to test in such a way. RDTs, although inexpensive, suffer from low sensitivity due to technical issues. In this article, we propose saliva as a noninvasive source for RT-PCR samples and evaluate various specimen types at different times after infection for the best possible output from COVID-19 rapid tests.
Keywords: COVID-19; RT-PCR; SARS-CoV-2; rapid diagnostic tests; saliva; salivary specimen.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

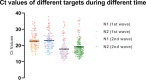
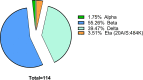
Similar articles
-
Diagnostic Performance Assessment of Saliva RT-PCR and Nasopharyngeal Antigen for the Detection of SARS-CoV-2 in Peru.Microbiol Spectr. 2022 Aug 31;10(4):e0086122. doi: 10.1128/spectrum.00861-22. Epub 2022 Jul 18. Microbiol Spectr. 2022. PMID: 35867471 Free PMC article.
-
Sample collection and transport strategies to enhance yield, accessibility, and biosafety of COVID-19 RT-PCR testing.J Med Microbiol. 2021 Sep;70(9):001380. doi: 10.1099/jmm.0.001380. J Med Microbiol. 2021. PMID: 34486972 Free PMC article.
-
Clinical Performance of Direct RT-PCR Testing of Raw Saliva for Detection of SARS-CoV-2 in Symptomatic and Asymptomatic Individuals.Microbiol Spectr. 2022 Dec 21;10(6):e0222922. doi: 10.1128/spectrum.02229-22. Epub 2022 Nov 21. Microbiol Spectr. 2022. PMID: 36409097 Free PMC article.
-
Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: a systematic review and meta-analysis.Lancet Infect Dis. 2021 Sep;21(9):1233-1245. doi: 10.1016/S1473-3099(21)00146-8. Epub 2021 Apr 12. Lancet Infect Dis. 2021. PMID: 33857405 Free PMC article.
-
Alternative clinical specimens for the detection of SARS-CoV-2: A rapid review.Rev Med Virol. 2021 Jul;31(4):e2185. doi: 10.1002/rmv.2185. Epub 2020 Oct 22. Rev Med Virol. 2021. PMID: 33091200 Review.
Cited by
-
Treating COVID-19 with Medicinal Plants: Is It Even Conceivable? A Comprehensive Review.Viruses. 2024 Feb 20;16(3):320. doi: 10.3390/v16030320. Viruses. 2024. PMID: 38543686 Free PMC article. Review.
-
Comprehensive Analysis of SARS-CoV-2 Dynamics in Bangladesh: Infection Trends and Variants (2020-2023).Viruses. 2024 Aug 7;16(8):1263. doi: 10.3390/v16081263. Viruses. 2024. PMID: 39205237 Free PMC article.
-
Rapid, sensitive, and specific detection of SARS-CoV-2 in nasopharyngeal swab samples of suspected patients using a novel one-step loop-mediated isothermal amplification (one-step LAMP) technique.BMC Microbiol. 2023 Mar 7;23(1):63. doi: 10.1186/s12866-023-02806-z. BMC Microbiol. 2023. PMID: 36882699 Free PMC article.
-
Performance of self-performed SARS-CoV-2 rapid antigen test: a systematic review and meta-analysis.Front Public Health. 2024 Oct 18;12:1402949. doi: 10.3389/fpubh.2024.1402949. eCollection 2024. Front Public Health. 2024. PMID: 39494084 Free PMC article.
-
Rapid Antigen Tests during the COVID-19 Era in Korea and Their Implementation as a Detection Tool for Other Infectious Diseases.Bioengineering (Basel). 2023 Mar 3;10(3):322. doi: 10.3390/bioengineering10030322. Bioengineering (Basel). 2023. PMID: 36978713 Free PMC article. Review.
References
-
- World Health Organization. 2022. WHO coronavirus (COVID-19) dashboard. https://covid19.who.int/. Accessed 10 May 2022.
-
- Jayk Bernal A, Gomes da Silva MM, Musungaie DB, Kovalchuk E, Gonzalez A, Delos Reyes V, Martín-Quirós A, Caraco Y, Williams-Diaz A, Brown ML, Du J, Pedley A, Assaid C, Strizki J, Grobler JA, Shamsuddin HH, Tipping R, Wan H, Paschke A, Butterton JR, Johnson MG, De Anda C, MOVe-OUT Study Group . 2022. Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. N Engl J Med 386:509–520. doi:10.1056/NEJMoa2116044. - DOI - PMC - PubMed
-
- Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, Lopez de Castilla D, Finberg RW, Dierberg K, Tapson V, Hsieh L, Patterson TF, Paredes R, Sweeney DA, Short WR, Touloumi G, Lye DC, Ohmagari N, Oh M-D, Ruiz-Palacios GM, Benfield T, Fätkenheuer G, Kortepeter MG, Atmar RL, Creech CB, Lundgren J, Babiker AG, Pett S, Neaton JD, Burgess TH, Bonnett T, Green M, Makowski M, Osinusi A, Nayak S, Lane HC, ACTT-1 Study Group Members . 2020. Remdesivir for the treatment of Covid-19—final report. N Engl J Med 383:1813–1826. doi:10.1056/NEJMoa2007764. - DOI - PMC - PubMed
-
- Colson P, Delerce J, Burel E, Dahan J, Jouffret A, Fenollar F, Yahi N, Fantini J, La Scola B, Raoult D. 2022. Emergence in southern France of a new SARS-CoV-2 variant harbouring both N501Y and E484K substitutions in the spike protein. Arch Virol 167:1185–1190. doi:10.1007/s00705-022-05385-y. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

