Cec4-Derived Peptide Inhibits Planktonic and Biofilm-Associated Methicillin Resistant Staphylococcus epidermidis
- PMID: 36453944
- PMCID: PMC9769716
- DOI: 10.1128/spectrum.02409-22
Cec4-Derived Peptide Inhibits Planktonic and Biofilm-Associated Methicillin Resistant Staphylococcus epidermidis
Abstract
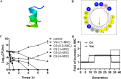
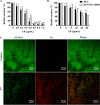
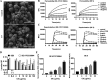
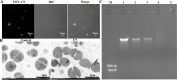
Staphylococcus epidermidis is part of the normal microbiota that colonizes the skin and mucosal surfaces of human beings. Previous studies suggested that S. epidermidis possessed low virulence, but recent studies confirmed that it can acquire high virulence from Staphylococcus aureus and with the increasing detection of methicillin-resistant S. epidermidis. It has become a major pathogen of graft-associated and hospital-acquired infections. In previous studies, we modified the antimicrobial peptide Cec4 (41 amino acids) and obtained the derived peptide C9 (16 amino acids) showing better antimicrobial activity against S. epidermidis with an MIC value of 8 μg/mL. The peptide has rapid bactericidal activity without detectable high-level resistance, showing certain inhibition and eradication ability on S. epidermidis biofilms. The damage of cell membrane structures by C9 was observed by scanning emission microscopy (SEM) and transmission electron microscopy (TEM). In addition, C9 altered the S. epidermidis cell membrane permeability, depolarization levels, fluidity, and reactive oxygen species (ROS) accumulation and possessed the ability to bind genomic DNA. Analysis of the transcriptional profiles of C9-treated cells revealed changes in genes involved in cell wall and ribosome biosynthesis, membrane protein transport, oxidative stress, and DNA transcription regulation. At the same time, the median lethal dose of C9 in mice was more than 128 mg/kg, and the intraperitoneal administration of 64 mg/kg was less toxic to the liver and kidneys of mice. Furthermore, C9 also showed a certain therapeutic effect on the mouse bacteremia model. In conclusion, C9 may be a candidate drug against S. epidermidis, which has the potential to be further developed as an antibacterial therapeutic agent. IMPORTANCE S. epidermidis is one of the most important pathogens of graft-related infection and hospital-acquired infection. The growing problem of antibiotic resistance, as well as the emergence of bacterial pathogenicity, highlights the need for antimicrobials with new modes of action. Antimicrobial peptides have been extensively studied over the past 30 years as ideal alternatives to antibiotics, and we report here that the derived peptide C9 is characterized by rapid bactericidal and antibiofilm activity, avoiding the development of resistance by acting on multiple nonspecific targets of the cell membrane or cell components. In addition, it has therapeutic potential against S. epidermidis infection in vivo. This study provides a rationale for the further development and application of C9 as an effective candidate antibiotic.
Keywords: S. epidermidis; antibacterial activity; antibacterial mechanism; antimicrobial peptide.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
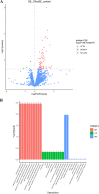

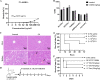
References
-
- Du X, Larsen J, Li M, Walter A, Slavetinsky C, Both A, Sanchez Carballo PM, Stegger M, Lehmann E, Liu Y, Liu J, Slavetinsky J, Duda KA, Krismer B, Heilbronner S, Weidenmaier C, Mayer C, Rohde H, Winstel V, Peschel A. 2021. Staphylococcus epidermidis clones express Staphylococcus aureus-type wall teichoic acid to shift from a commensal to pathogen lifestyle. Nat Microbiol 6:757–768. doi: 10.1038/s41564-021-00913-z. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

